Description
The DepleteX® Single Cell RNA Boost Kit employs a CRISPR-based method to selectively remove unwanted reads allowing you to cut through the noise with minimal impact on your workflow and maximum confidence in your results.
For research use only. Not for use in diagnostic procedures.
Overview
Enhance usable single-cell RNA-seq data using DepleteX ® Single Cell RNA Boost Kit
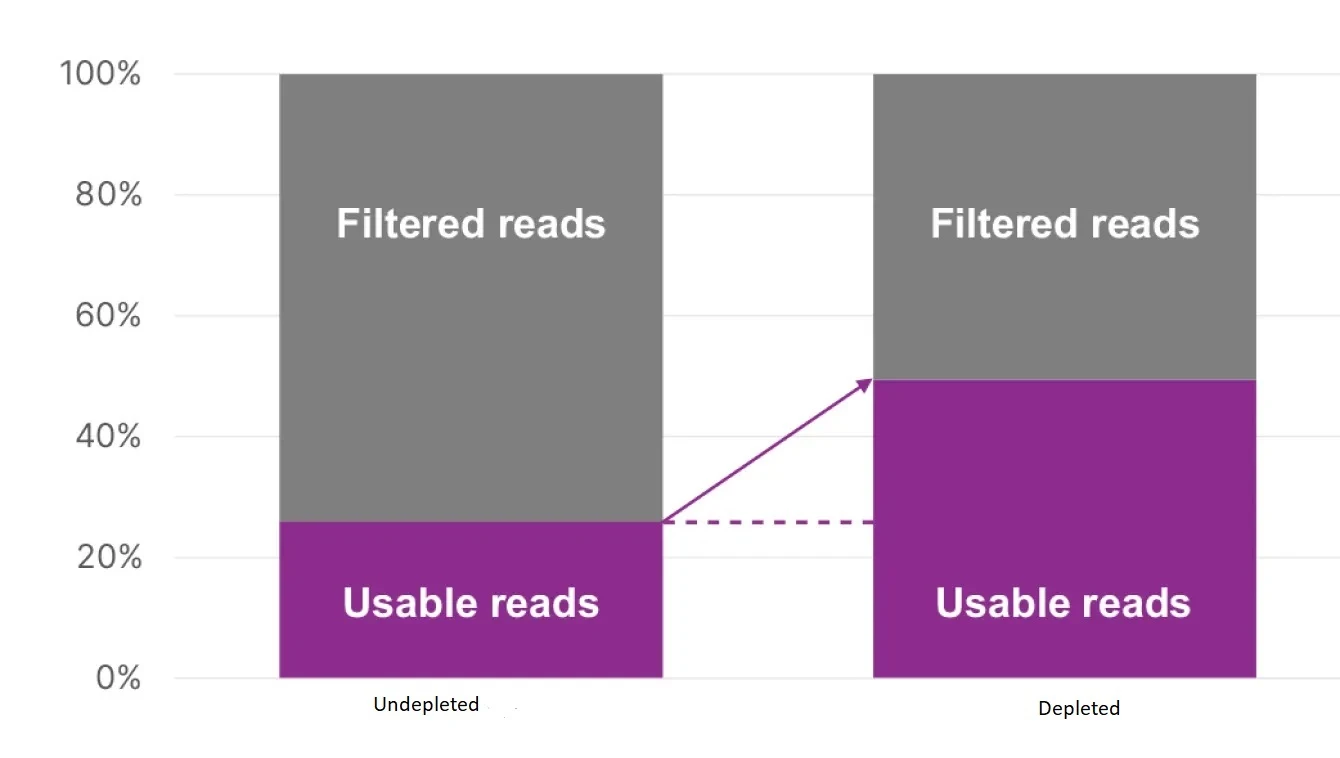
DepleteX ® efficiently removes abundant and uninformative fragments before sequencing, reallocating 50% of sequencing clusters to informative reads that would typically be filtered out during secondary analysis. This kit utilizes Cas9 depletion with an optimized guide set to target ribosomal and mitochondrial mRNA, non-transcriptomic reads, and highly expressed non-variable genes. Additionally, the kit provides a separate tube for non-variable gene depletion, which can be included or excluded as required.
The DepleteX® Single Cell RNA Boost Kit empowers you to reduce noise and optimize usable data in your single-cell RNA sequencing.
Key highlights:
- Enhance secondary analysis with ~50% more useful reads
- Verified for both short-read and long-read sequencing.
- Efficiently removes ribosomal, mitochondrial, non-transcriptomic reads, and optionally non-variable genes from single-cell libraries
- Streamlined 3-step protocol
- Tested with 10X Genomics Single Cell Sequencing and other scRNA sequencing workflows
Are you using blood as a sample type for your single cell sequencing experiments? Learn how you can also remove unwanted globin transcripts from your data.
Additional product information
Reducing costs associated with single cell sequencing.
Implementing single-cell sequencing can be costly. However, the DepleteX® Single Cell RNA Boost Kit provides cost savings by efficiently eliminating abundant and uninformative fragments before sequencing, reallocating 50% of sequencing clusters to informative reads that would typically be filtered out during secondary analysis.

Figure 1: Double the reads mapped to the transcriptome with the DepleteX® Single Cell RNA Boost Kit.
Reduction of single cell sequencing data complexity.
Single-cell sequencing generates large and complex datasets. Analyzing these data requires robust computational methods and expertise. The sensitivity of scRNA sequencing can lead to high levels of technical noise. The DepleteX® Single Cell RNA Boost Kit empowers you to reduce noise and optimize usable data in your single-cell RNA sequencing. Figure 2 illustrates a 1.5x improvement in detection after depletion with this kit.
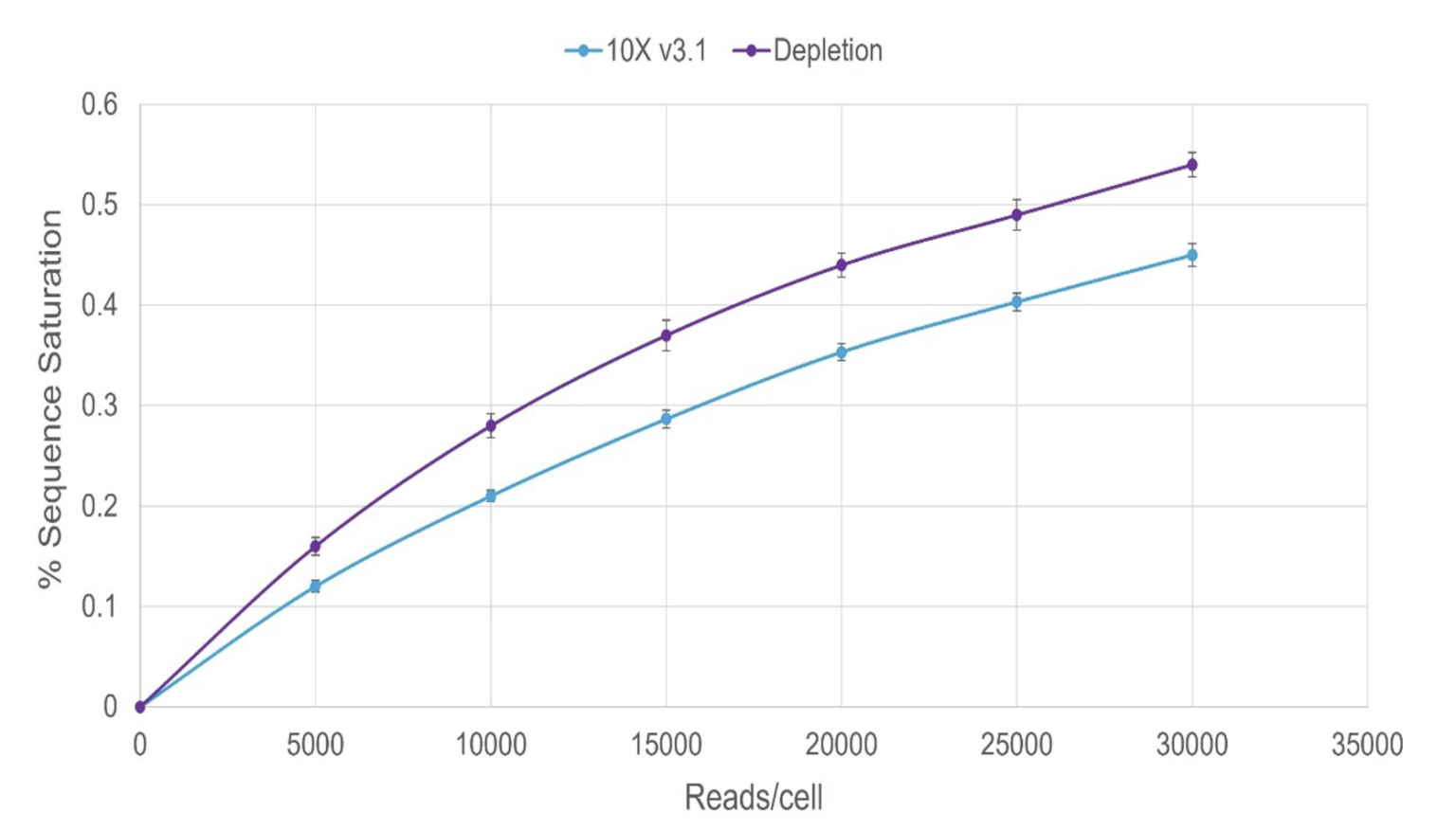
Figure 2: 1.5x improvement in detection with depletion
FAQs
Q: Can I use the DepleteX® Single Cell RNA Boost Kit on mouse samples?
A: Yes, but the depletion rate is lower since the guides are designed for human samples.
Q: Does the DepleteX® Single Cell RNA Boost Kit use all of the cDNA from my 10X prep?
A: No, only 1 tube of the available cDNA from a given 10X prep is used in general. The customer is free to customize this as needed to hit the 100-150 ng input recommendation.
Q: Is this kit compatible with the 5’ Chromium assay (10X Genomics), the TotalSeq™ reagents and Visium™ spatial (10X Genomics)?
A: Yes, it is compatible with all these workflows. Our depletion panel is designed to tile across the full length of transcripts.
Q: What is the sequencing depth recommendation for the depleted condition?
A: general, single cell users use between 25K – 50K reads/cell depending on their application. For the depleted condition, users can choose to sequence 12.5K – 25K reads/cell to see the same information or sequence at the same depth to see more information.
Q: Can I still use mitochondrial reads to separate “intact” cell from “compromised” cells?
A: Mitochondrial reads are targeted by the DepleteX® Single Cell RNA Boost Kit but removal is not complete. They can be used to separate intact from compromised cells. Instead of setting an arbitrary 10% threshold, alternative approaches are described in the Python and R guides provided.
Specifications
| Automation Compatible |
No
|
|---|---|
| Product Group |
Single Cell Depletion
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
24 rxns
|
References
- Currenti, J., Qiao, L., Pai, R., Gupta, S., Khyriem, C., Wise, K., Sun, X., Armstrong, J., Crane, J., Pathak, S., Yang, B., George, J., Plummer, J., Martelotto, L., & Sharma, A. (2022). STOmics-GenX: CRISPR-based approach to improve cell identity-specific gene detection from spatially resolved transcriptomics. bioRxiv. Advance online publication.
- Pandey, A. C., Bezney, J., DeAscanis, D., Kirsch, E., Ahmed, F., Crinklaw, A., Choudhary, K. S., Mandala, T., Deason, J., Hamdi, J., Siddique, A., Ranganathan, S., Ordoukhanian, P., Brown, K., Armstrong, J., Head, S., & Topol, E. J. (2022). A CRISPR/Cas9-based enhancement of high-throughput single-cell transcriptomics. bioRxiv. Advance online publication.


