Description
The DepleteX® Nasopharyngeal Microbial RNA Boost Kit selectively removes highly expressed transcripts in vivo before sequencing. This process redistributes sequencing clusters toward microbial content and pathogens, enabling maximum discovery.
For research use only. Not for use in diagnostic procedures.
Overview
Enhance microbial discovery by reducing abundant human reads.
Nasopharyngeal RNA samples often exhibit low quality and quantity due to dominance by host and commensal microbial RNA. This combination of factors— low input, poor quality RNA, and high host contamination—reduces sensitivity in metagenomic next-generation sequencing (mNGS) and hinders the detection of low-expressing microbes and novel pathogens.
Key highlights of the DepleteX® Nasopharyngeal Microbial RNA Boost Kit:
- Efficient human RNA depletion from nasopharyngeal samples
- Enhanced pathogen detection in challenging nasopharyngeal samples with significant human RNA presence.
- Facilitates virus detection at high Cq values (up to 30) using 4 million reads.
Additional Product Information:
The DepleteX® Nasopharyngeal Microbial RNA Boost Kit is used during the RNA-seq library prep workflow starting with nasopharyngeal samples after the PCR step and then Post-depletion PCR amplification ensures that only uncleaved fragments, i.e., those containing sequences of interest, are amplified further. A final bead-based cleanup step ensures that the library is ready for sequencing. This kit is compatible with most RNA-seq workflows; however, we recommend using it with the NEXTFLEX® Rapid Directional RNA-seq kit 2.0.
Additional product information
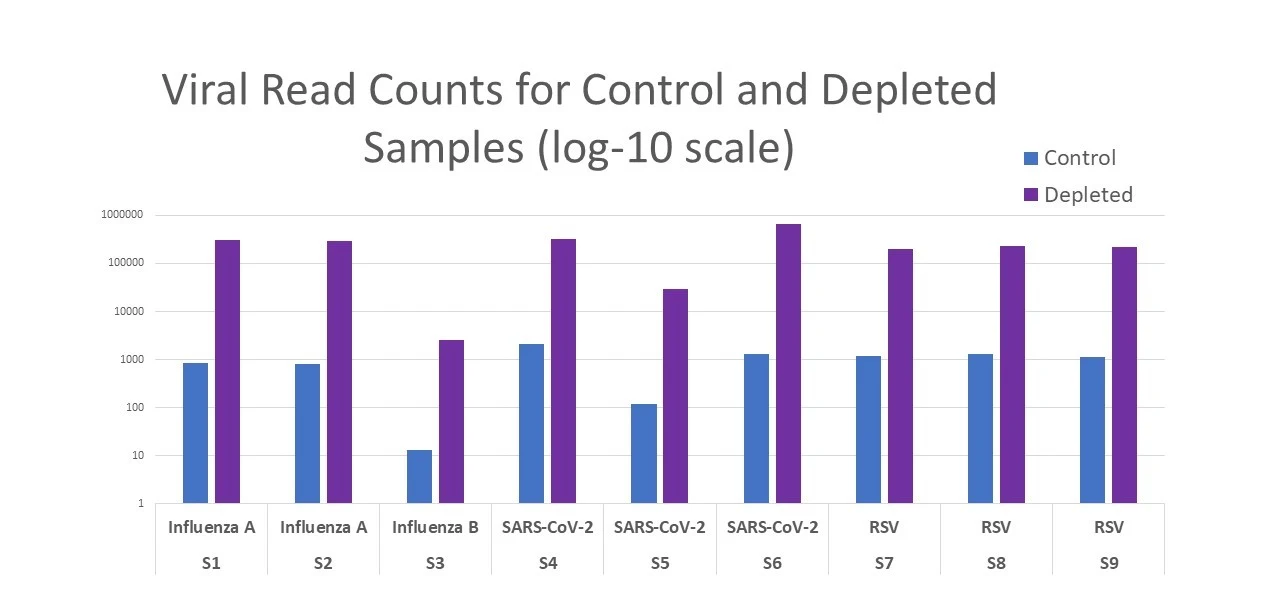
Figure 1 illustrates the enhanced pathogen detection capabilities of the kit achieved by efficient depletion of human and bacterial rRNA and overabundant human mRNA.

Figure 1. Average of 265-fold change in viral read count between control and depleted samples.
Specifications
| Automation Compatible |
No
|
|---|---|
| Product Group |
Ribodepletion
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
24 rxns
|
References
- Brown, K., Chan, A. P., Siddique, A., Desplat, Y., Choi, Y., Ranganathan, S., Choudhary, K. S., Diaz, J., Bezney, J., DeAscanis, D., George, Z., Wong, S., Selleck, W., Bowers, J., Zismann, V., Reining, L., Highlander, S., Hakak, Y., & Armstrong, J. R. (2022). A Universal Day Zero Infectious Disease Testing Strategy Leveraging CRISPR-based Sample Depletion and Metagenomic Sequencing. Open Forum Infectious Diseases, 9(Supplement_2), ofac492.5801.
- Cerón, S., Clemons, N. C., von Bredow, B., & Yang, S. (2023). Application of CRISPR-Based Human and Bacterial Ribosomal RNA Depletion for SARS-CoV-2 Shotgun Metagenomic Sequencing. American Journal of Clinical Pathology, 159(2), 111–1151.


