
Description
The NEXTFLEX™ Rapid Directional RNA-seq library prep kit 2.0 produces libraries for Illumina® and Element Biosciences® sequencing instruments with high coverage uniformity, low duplication rates, strand specificity and minimal rRNA contamination when used with the NEXTFLEX Poly(A) Beads 2.0 (10 ng – 5 μg) or NEXTFLEX RiboNaut™ rRNA depletion kit (human, mouse, rat) (5 ng – 1 μg).
For research use only. Not for use in diagnostic procedures.
Overview
- High coverage uniformity with low duplication rate
- Optimized for use with 5 ng – 5 µg total RNA with reverse transcriptase and cleanup/size selection beads included
- Simple protocol validated with NEXTFLEX® poly(A) beads 2.0 and NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat)
- Designed to work with NEXTFLEX® RNA-Seq 2.0 Unique Dual Index barcodes that allow a wide range of multiplexing (2 up to 1,526 samples in one run)
- Up to 96 UDI-UMI Barcodes now available to improve gene expression analysis by removing PCR duplicates.
- Automated on the Sciclone® G3 NGSx and Zephyr® G3 NGS workstations
- Compatible with Illumina® and Element Biosciences® sequencing platforms
The NEXTFLEX® Rapid Directional RNA-seq kit 2.0 includes reverse transcriptase, necessary library preparation reagents, and cleanup/size selection beads optimized to ensure reliable performance. The kit involves a simple library preparation protocol that has been tested with the NEXTFLEX® poly(A) beads 2.0 and NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) to accommodate total RNA as input. The NEXTFLEX® rapid directional RNA-seq kit 2.0 is designed to be used with NEXTFLEX® RNA-Seq 2.0 Unique Dual Index Barcodes (6.25 μM), which are color-balanced and have undergone proprietary purity QC metrics to generate reliable sequencing results for every sample.
Additional product information

Performance of Poly(A)-selected Libraries
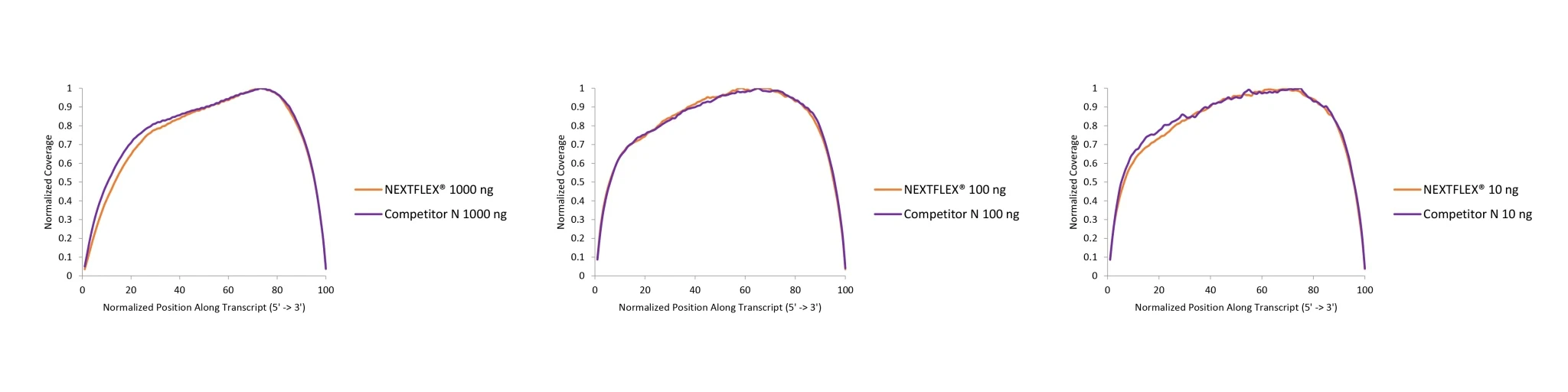
Figure 1. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrates even coverage along transcripts compared to the Competitor N kit.
Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX Poly(A) beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq® sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference using bowtie2. The coverage along transcripts was calculated using the BBMap pileup tool.
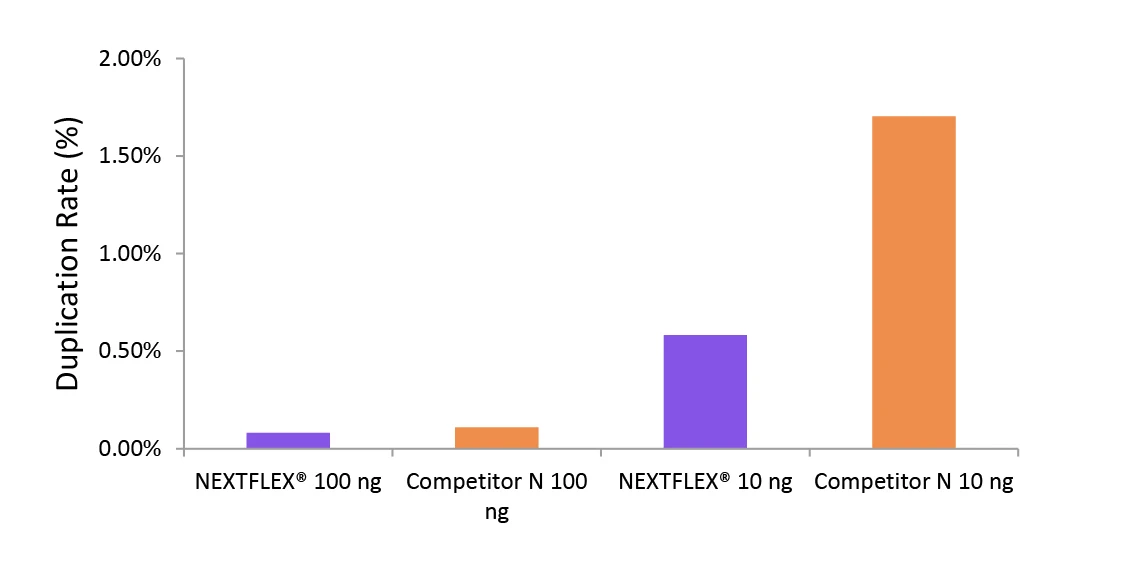
Figure 2. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrate low duplication rate compared to the Competitor N kit.
Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent ®#740000) using the NEXTFLEX Poly(A) beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference using bowtie2, and randomly downsampled to 100k reads. Duplication rate was calculated using the fastp all-in-one FASTQ preprocessor.
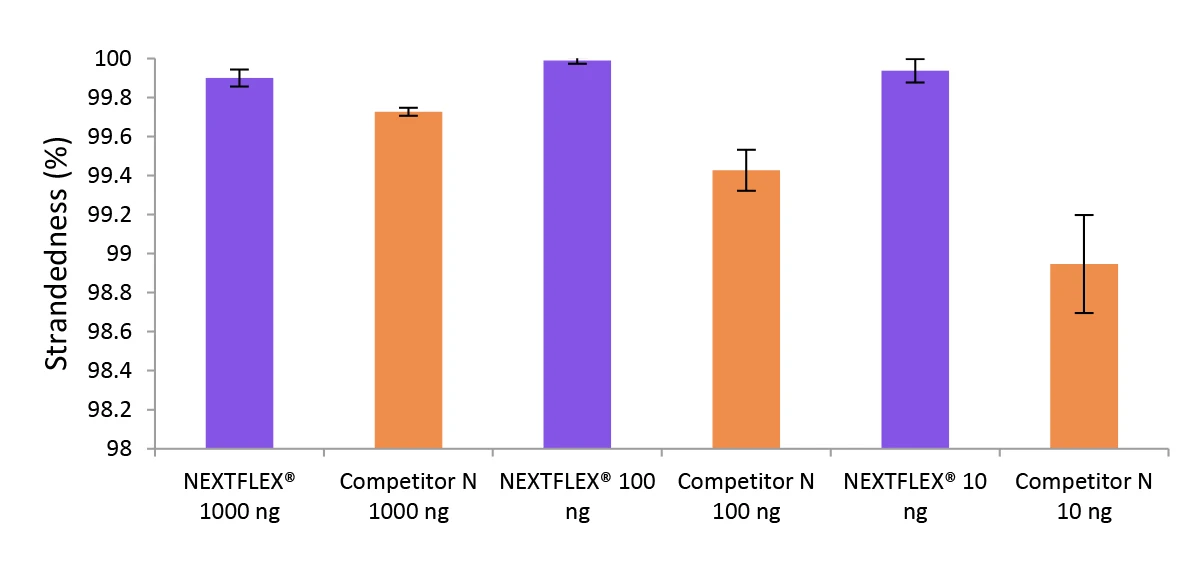
Figure 3. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrates comparable directionality to the Competitor N kit.
Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) containing ERCC RNA Spike-In mix (Thermo Fisher Scientific #4456740) using the NEXTFLEX Poly(A) Beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt and mapped to the ERCC92 reference using bowtie2. Strandedness was calculated using SAMtools.
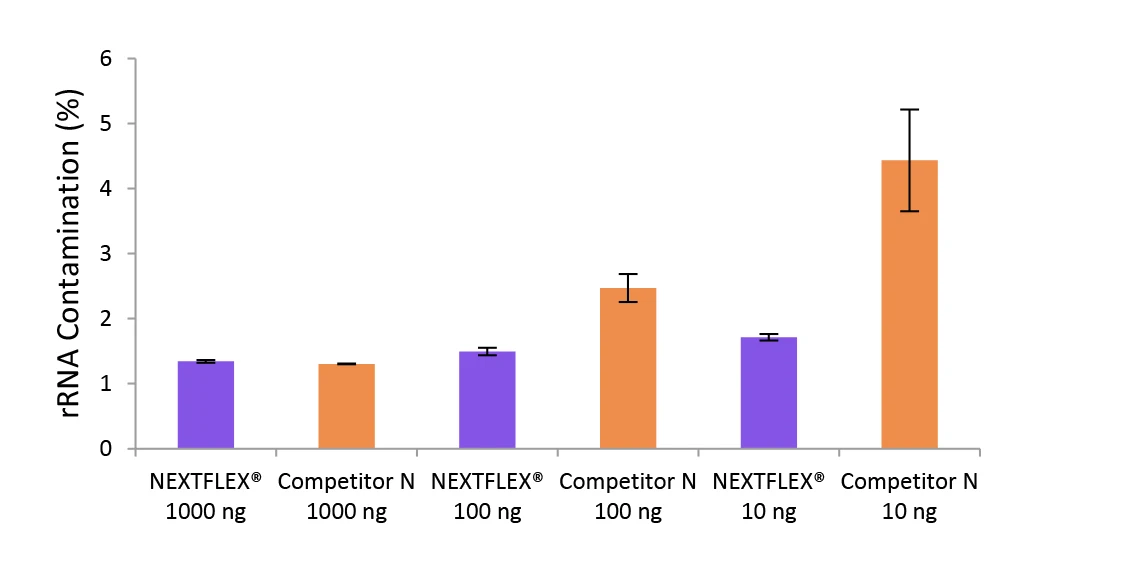
Figure 4. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 delivers libraries containing low levels of rRNA contamination than the Competitor N kit.
Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX Poly(A) Beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using paired-end mode (2×76 bp). The reads were trimmed using cutadapt and the percent of rRNA was determined by using bowtie2 to map reads to human rRNA. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrated superior removal of 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species compared to the Competitor N kit.
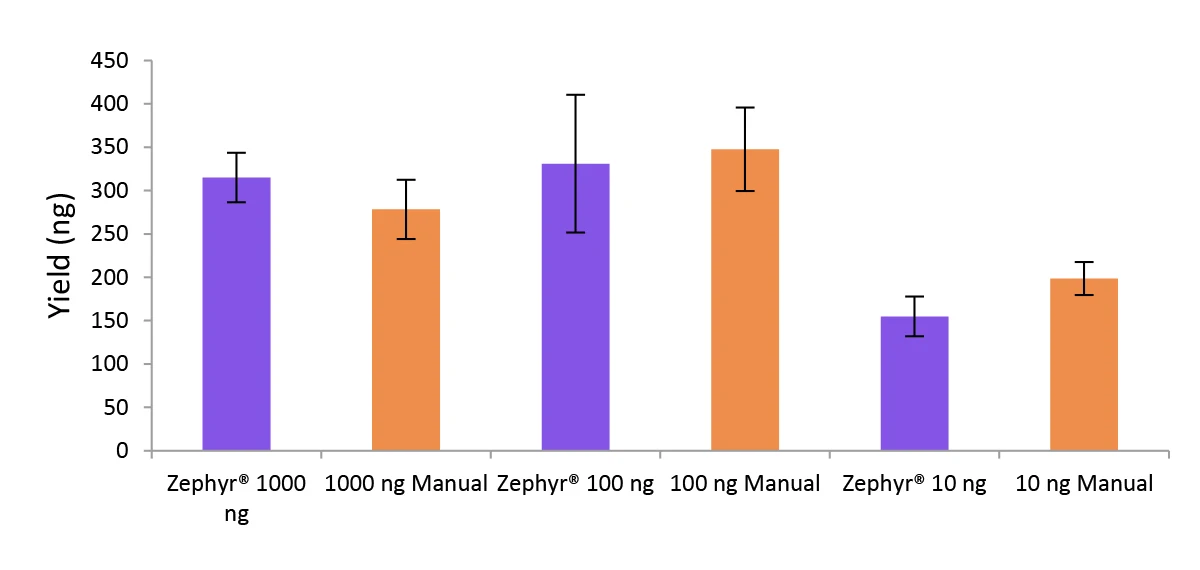
Figure 5. Libraries prepared using the Zephyr G3 NGS workstation and manually deliver comparable yields using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0.
Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX Poly(A) Beads 2.0. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0. Final library concentrations were quantified using the Qubit® 2.0 fluorometer (Thermo Fisher® Scientific #Q32866).
Performance of rRNA-depleted Libraries
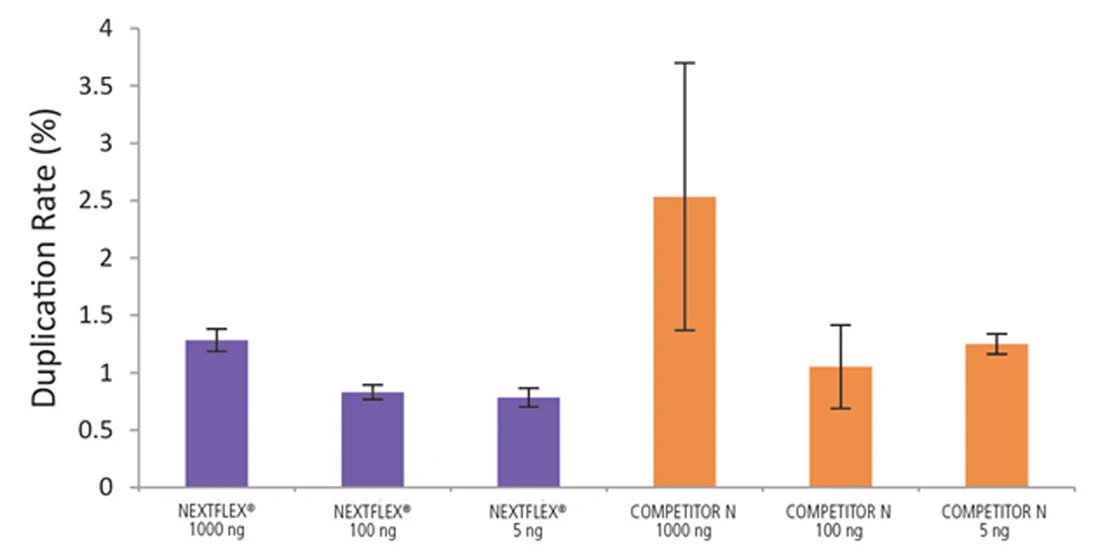
Figure 6. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrate low duplication rates compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference transcriptome using bowtie2, and randomly downsampled to 28k reads. Duplication rate was calculated using the fastp all-in-one FASTQ preprocessor.
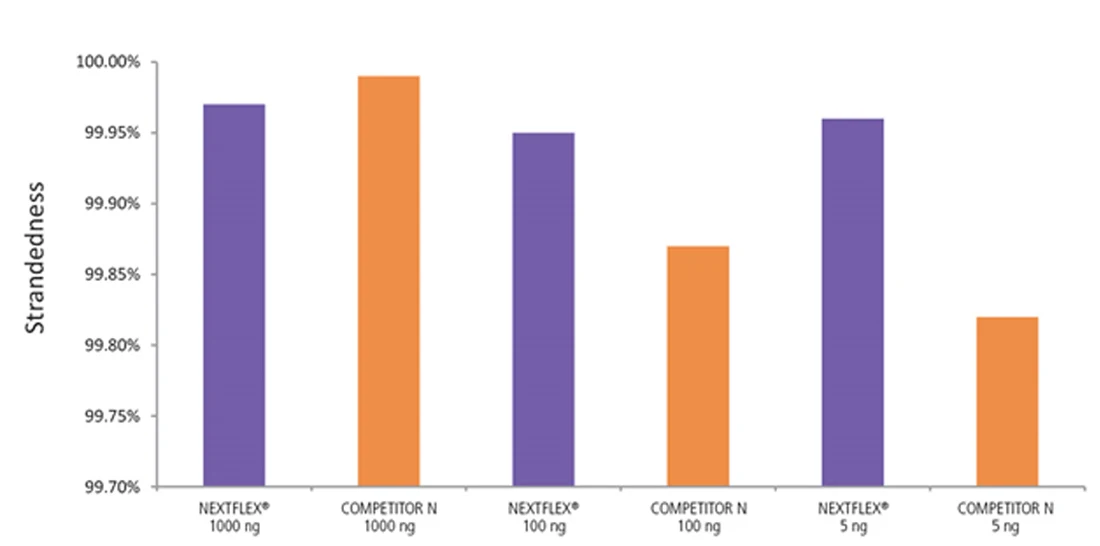
Figure 7. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrates comparable directionality relative to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference transcriptome using bowtie2. Reads from respective samples were combined and downsampled for a total of 800k reads each. Strandedness was calculated using the fastp all-in-one FASTQ preprocessor.
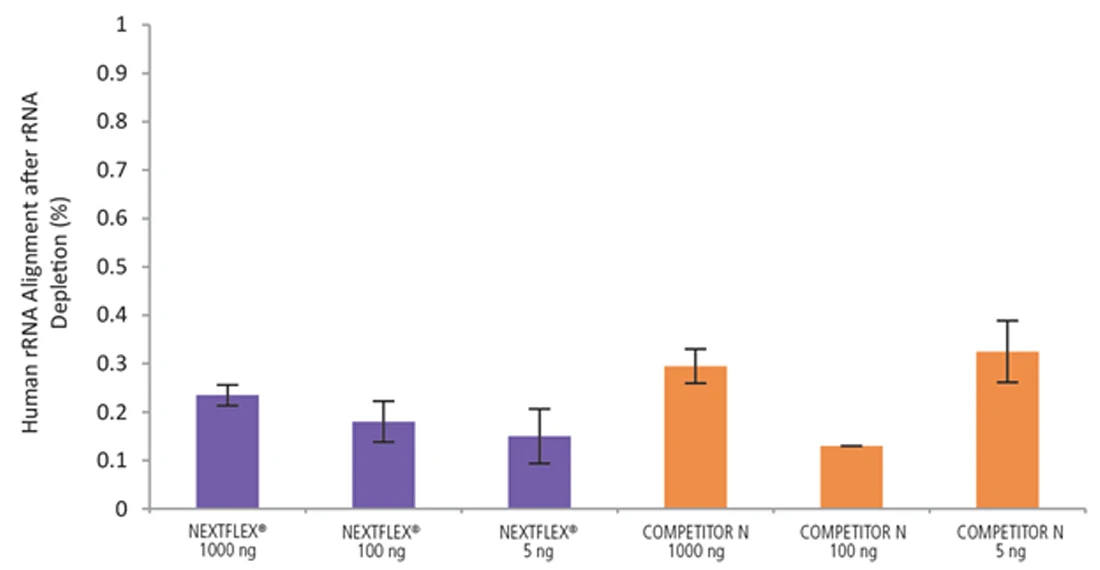
Figure 8. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 delivers libraries containing low levels of rRNA contamination compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). The reads were trimmed using cutadapt and the percent of rRNA was determined by using bowtie2 to map reads to human rRNA. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrated superior removal of 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species compared to the Competitor N kit.
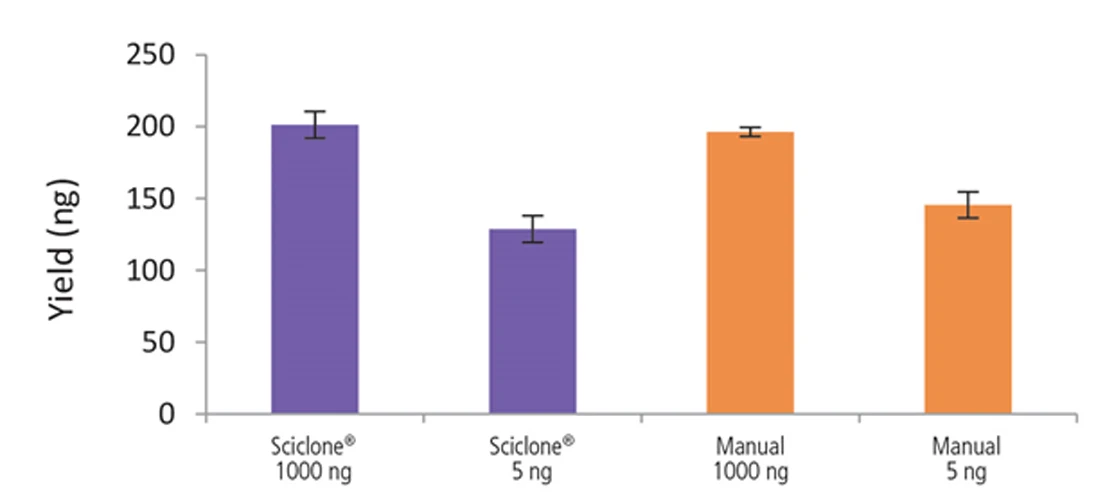
Figure 9. Libraries prepared using the Sciclone G3 NGSx workstation and manually deliver comparable yields using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0. Final library concentrations were quantified using the Qubit 2.0 fluorometer (Thermo Fisher Scientific #Q32866).
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Brand |
NEXTFLEX
|
| Product Group |
RNA-seq
|
| Shipping Conditions |
Dual Temperature
|
| Type |
RNA-seq
|
| Unit Size |
48 rxns
|
Citations
- Bond, D.M., Ortega-Recalde, O., Laird, M.K. et al. The admixed brushtail possum genome reveals invasion history in New Zealand and novel imprinted genes. Nat Commun 14, 6364 (2023). doi.org/10.1038/s41467-023-41784-8.
- Gaonkar, C.C., Campbell, L. De novo transcriptome assembly and gene annotation for the toxic dinoflagellate Dinophysis. Sci Data 10, 345 (2023). doi.org/10.1038/s41597-023-02250-8.
- Laudadio, I., Carissimi, C., Scafa, N. et al. Characterization of patient-derived intestinal organoids for modelling fibrosis in Inflammatory Bowel Disease. Inflamm. Res. 73, 1359–1370 (2024). https://doi.org/10.1007/s00011-024-01901-9.
- Manukjan, N., Chau, S., Caiment, F. et al. Wnt7a Decreases Brain Endothelial Barrier Function Via β-Catenin Activation. Mol Neurobiol 61, 4854–4867 (2024). doi.org/10.1007/s12035-023-03872-0
- Mulroney, T.E., Pöyry, T., Yam-Puc, J.C. et al. N1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting. Nature 625, 189–194 (2024). doi.org/10.1038/s41586-023-06800-3
- Smith, C. H., Mejia-Trujillo, R., Breton, S., Pinto, B. J., Kirkpatrick, M., & Havird, J. C. (2023). Mitonuclear Sex Determination? Empirical Evidence from Bivalves. Molecular Biology and Evolution, 40(11), msad240.
- Tóvári, J.; Vári-Mező, D.; Surguta, S.E.; Ladányi, A.; Kigyós, A.; Cserepes, M. Evolving Acquired Vemurafenib Resistance in a BRAF V600E Mutant Melanoma PDTX Model to Reveal New Potential Targets. Cells 2023, 12, 1919. doi.org/10.3390/cells12141919.


