Description
Isolation of total DNA from stool samples in 96‑well plate format
| Selling unit | 96 Prep(s), 384 Prep(s) |
| Application | Isolation of DNA |
| Target | DNA |
| CE certified | No, research use only |
| Technology | Silica membrane technology |
| Brand | NucleoSpin |
| Format | 96-well plate |
| Handling | Vacuum |
| Lysate clarification | Vacuum, NucleoSpin Stool Filter Plate |
| Automated use | Yes |
| Application note available with | Eppendorf |
| Sample material | Stool |
| Fragment size | 50 bp–approx. 50 kbp |
| Typical yield | 3–15 μg (varies by sample and disruption device) |
| Theoretical binding capacity | 50 µg |
| Typical purity A260/A280 | 1.6–1.8 |
| Elution volume | 100−200 µL |
| Preparation time | 90 min/48 samples (automated use) |
| Typical downstream application | Metagenomics, NGS, qPCR |
| Storage temperature | 18–25 °C |
| Shelf life (from production) | 24 Month(s) |
| Hazardous material | Yes |
NucleoSpin 96 DNA Stool
NucleoSpin 96 DNA Stool was developed for the reliable, automation-friendly isolation of high quality DNA from human and animal stool samples. The kit combines a robust filtration step, significantly reducing the chance of column clogging, with an efficient DNA extraction procedure based on silica membrane technology.
Reliable performance and excellent yields
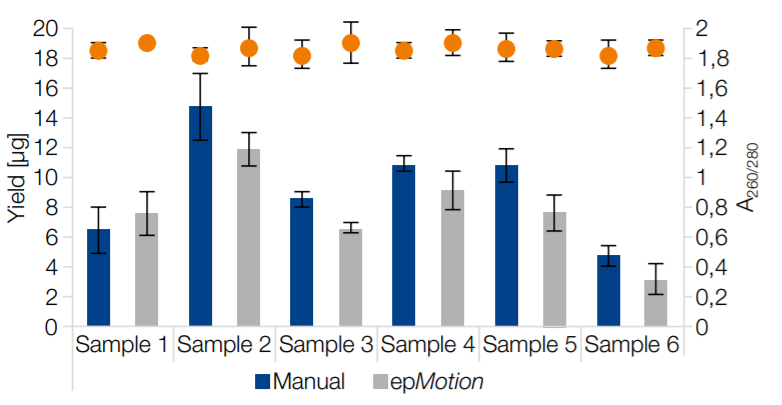
Comparable performance both manually and on epMotion 5075v
Six individual human stool samples were processed manually and on epMotion 5075v in quadruplicates. Yield (blue/grey bars) and purity (A260/280 nm, orange dots) were measured for all of the preparations. The manual and automated extractions delivered comparable yields with high purity (A260/280 > 1.7 in all samples)
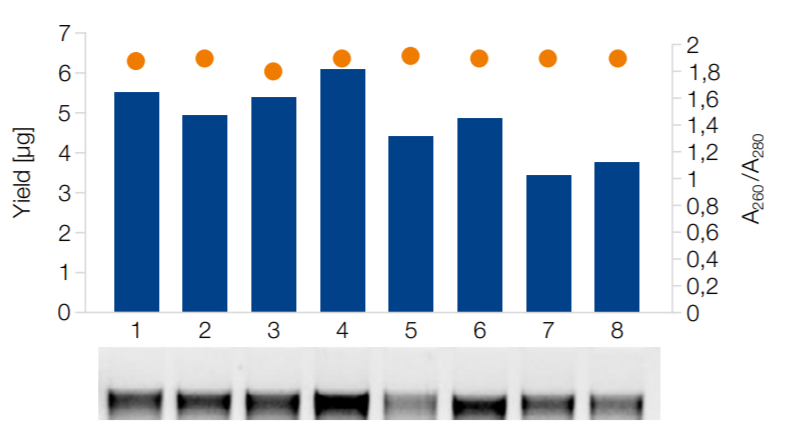
High quality DNA from rat stool samples
Eight individual rat stool samples were processed on epMotion 5075v. Yield (blue bars), purity (A260/280 nm, orange dots) and analysis of DNA on the gel indicate the reliably high quality of the extracted DNA.
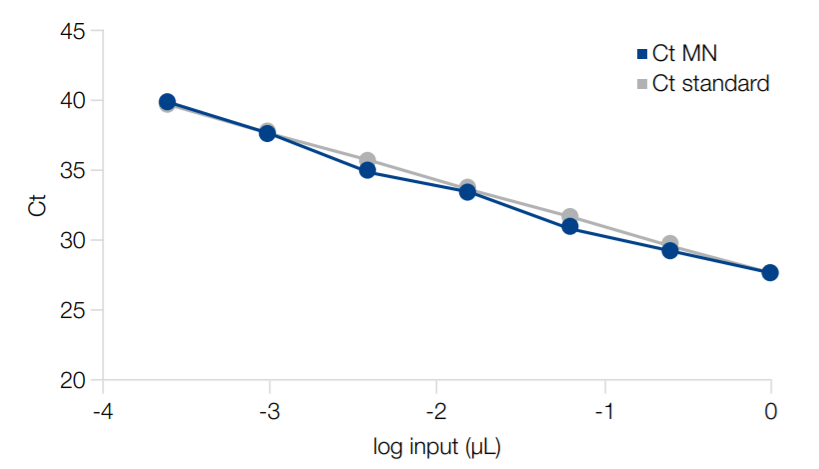
No downstream inhibition
DNA was purified from a human stool sample on epMotion 5075v using NucleoSpin 96 DNA Stool. A series of 1:4 dilutions, ranging from the undiluted eluate to a 4096fold dilution was analyzed by qPCR using an E.coli specific target and compared against a theoretical standard in a logarithmic plot. The slope of the regression curve (-3.4106) shows an excellent qPCR-performance without PCR inhibition.
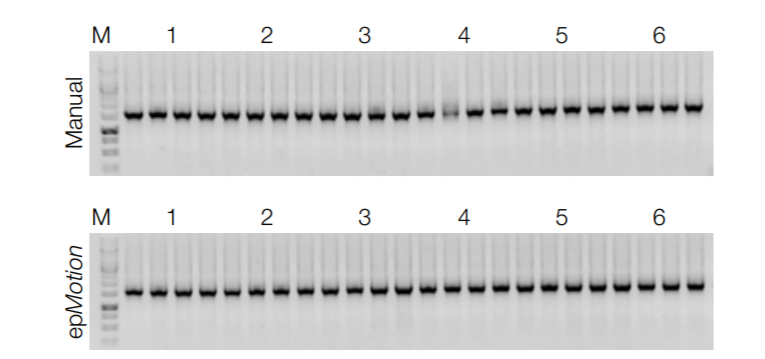
Consistently excellent PCR performance
Six individual human stool samples were processed manually and on epMotion 5075v in quadruplicates. From each eluate, 2,5 µL were used in a 25 µL endpoint PCR reaction, amplifying a 1,5 kb fragment of the bacterial 16SrRNA gene. Three µL from each PCR reaction were analyzed on a 1 % agarose/TAE gel. In each case, the samples were successfully amplified, providing a good indication for the robust performance of the DNA in down‑ stream applications.


