Description
Boost usable single cell RNA seq data with CRISPRclean.

Traditionally, single cell data processing incorporates certain filtering and normalization steps prior to cell clustering and downstream interpretation. Instead of removing those reads in-silico, CRISPRclean removes those reads in-vitro ahead of sequencing, redistributing 50% sequencing clusters to unique biologically relevant transcripts—allowing you to maximize UMI and gene sensitivity.
CRISPRclean leverages Cas9 depletion and a specifically designed guide set to remove reads that are filtered before secondary analysis. CRISPRclean Single Cell RNA Boost Kit gives you the ability to cut through the noise with minimal impact on your workflow, and maximum confidence in your single cell RNA seq results.
Highlights
- Gain a deeper view of expression profiles of individual cells
- 1.5x improvement in gene and UMI detection sensitivity
- Boost usable data by cutting wasted sequencing by 50%
- Depletes sequences not used for secondary analysis including unaligned reads, ribosomal, mitochondrial, and non-variable genes
*Data from PBMC’s, some variance expected with different cell types
| Specification | |
| Catalog | JUM-KIT1018 |
|---|---|
| Samples per kit | 24 |
| Assay time | 2 hours |
| Hands-on-time | 45 minutes |
| Input | Uses one of four Chromium cDNA aliquots per prep |
| Method | Single cell 3' gene expression libraries for 10x Genomics |
| Designed to deplete | Unaligned reads, ribosomal, mitochondrial, non-variable genes |
| Jumpcode validated | 10x Genomics Chromium Next GEM Single Cell 3' Reagent Kits v3.1, Visium Spatial Gene Expression libraries, and PacBio Iso-Seq SMRTbell Express Template Prep Kit 2.0 |
Simple 3-step protocol integrated into 10x Genomics® Chromium™ Next GEM Single Cell 3’ workflow

100% increase in reads mapped to the transcriptome with CRISPRclean Single Cell RNA Boost Kit.

1.5x improvement in detection sensitivity with depletion

Boost in detection sensitivity of genes and UMIs per cell for depleted PBMC samples compared to undepleted sample.
Gain a deeper view of expression profiles
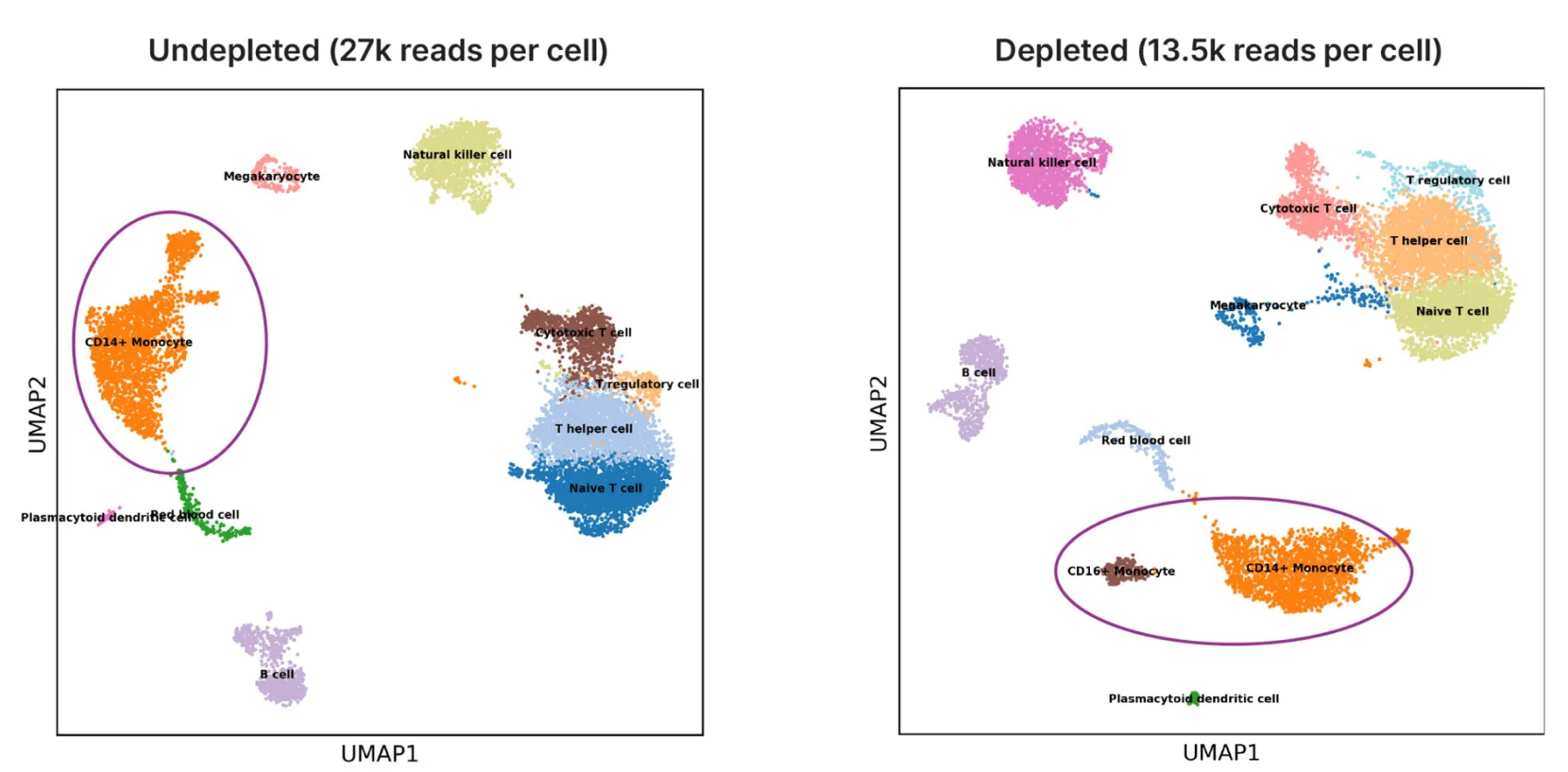 One additional cell type was identified in PBMC samples treated with CRISPRclean. UMAP plots of cell clusters with and without depletion using PBMC samples showed that depletion did not perturb cell type calls.
One additional cell type was identified in PBMC samples treated with CRISPRclean. UMAP plots of cell clusters with and without depletion using PBMC samples showed that depletion did not perturb cell type calls.


