Description
Plant total RNA is comprised of large amounts of undesired ribosomal RNA (rRNA) which accounts for up to ~97 % of all transcripts. Lexogen’s RiboCop rRNA Depletion Kits for Plants remove undesired cytoplasmic (5.8S, 18S and 25S), mitochondrial (5S, 18S and 26S) and plastid (4.5S, 5S, 16S and 23S) rRNA from intact as well as degraded input RNA. Ribo-depletion affords an unbiased view of the transcriptome, including non-coding and non-polyadenylated RNA species.
RiboCop for Plants is ideally suited for Next Generation Sequencing (NGS) approaches in combination with CORALL RNA-Seq V2 Library Preparation Kits. RiboCop Kits are also compatible with other random primed total RNA library prep kits.
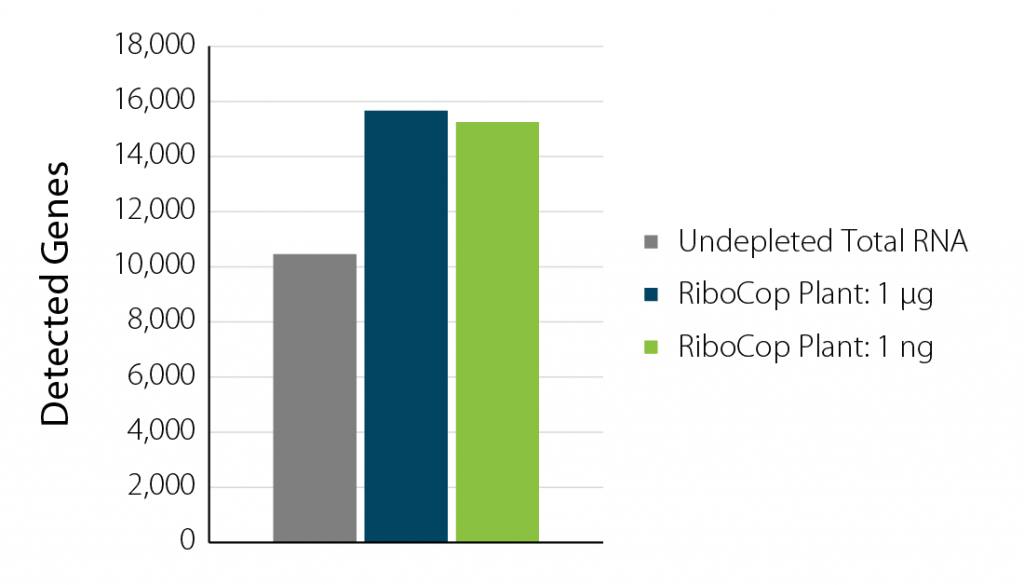
Figure 2 | Increased gene detection upon rRNA depletion. A. thaliana RNA samples were depleted with RiboCop for Plants using either 1 µg or 1 ng input RNA. Libraries were prepared using Lexogen’s CORALL RNA-Seq V2 Library Prep Kit and sequenced on Illumina NextSeq2000 (1×90 bp). Reads were mapped against the A. thaliana reference genome using STAR aligner and counted with FeatureCounts.
RiboCop for Plants Performance across Species
RiboCop enriches RNAs of interest across a range of different plant species (Fig. 3) and is suitable for depletion of various crops including corn (Zea mays), soybean (Glycine max), and others.
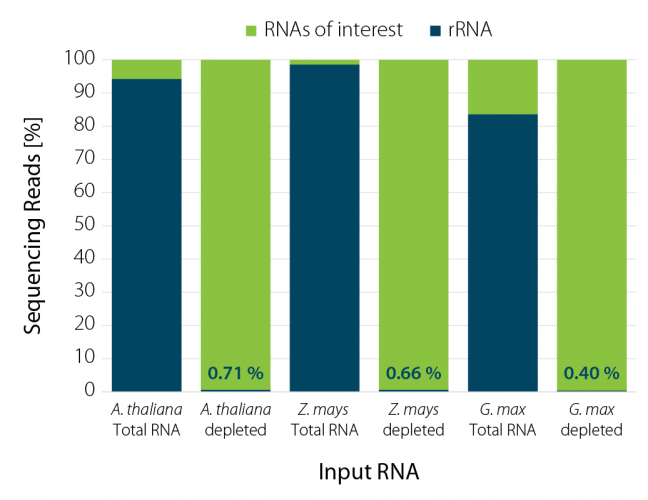
Figure 3 | RiboCop rRNA Depletion for Plants efficiently removes rRNA from A.thaliana, Z. mays, and G. max. NGS libraries were prepared using Lexogen’s CORALL RNA-Seq V2 Library Prep Kit. Successful depletion was monitored by NGS sequencing (NextSeq2000, 1×90 bp) and subsequent analysis of remaining rRNA reads from 5 ng untreated (Total RNA). 50 ng RNA from A. thaliana, Z. mays, and G. max were used for rRNA depletion prior to library generation. Reads were mapped to the respective reference genomes and the percentage of reads mapping to rRNA is plotted in blue.



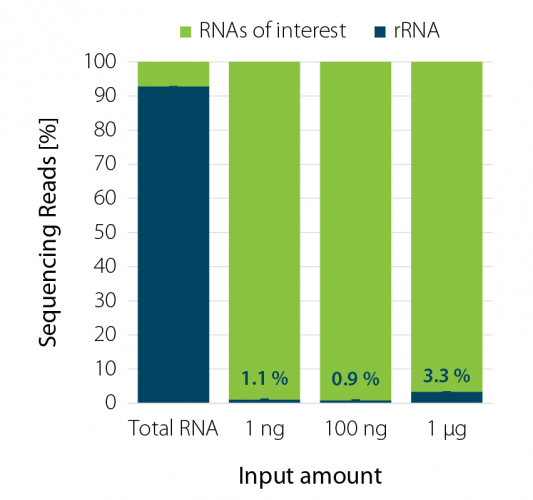 Figure 1 | RiboCop rRNA Depletion for Plants efficiently removes rRNA across a wide range of input amounts.
Figure 1 | RiboCop rRNA Depletion for Plants efficiently removes rRNA across a wide range of input amounts.