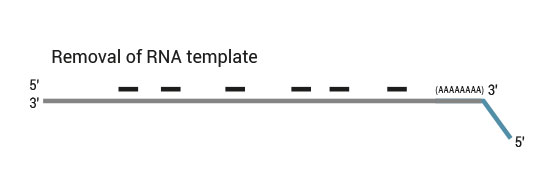
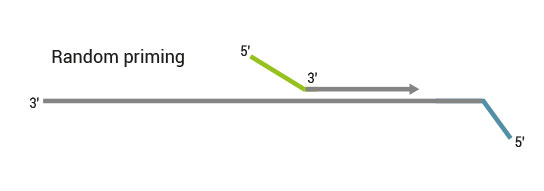
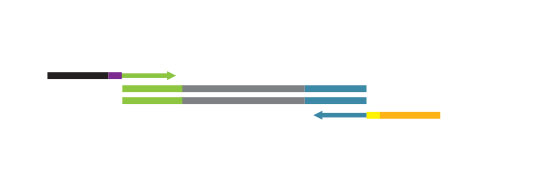
Description
QuantSeq with UDI V2 kits are provided with four essential components:
- a library generation module (same as the original QuantSeq FWD 015)
- a library amplification module
- a library purification module (magnetic beads)
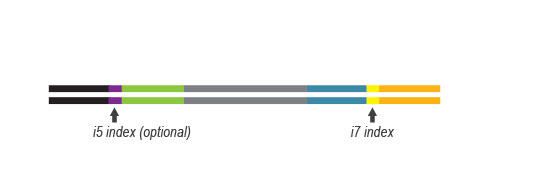
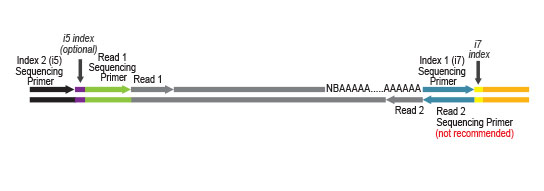
- a plate containing 24 or 96 Unique Dual Indices (UDI), packaged in single reactions
- for the 384-reaction kit: four plates containing 384 different UDI (sets A1 to A4)
UDI Set B1 is designed for Illumina chemistries of iSeq 100, MiniSeq, NextSeq 500 – 2000, HiSeq 3000 and 4000, as well as NovaSeq 6000 (v1.5 reagent kits).
Sets A1 to A4 can also be used for the above instruments, but sequences are reversed (this needs to be specified during data analysis).
Features and Application:
- Gene expression studies
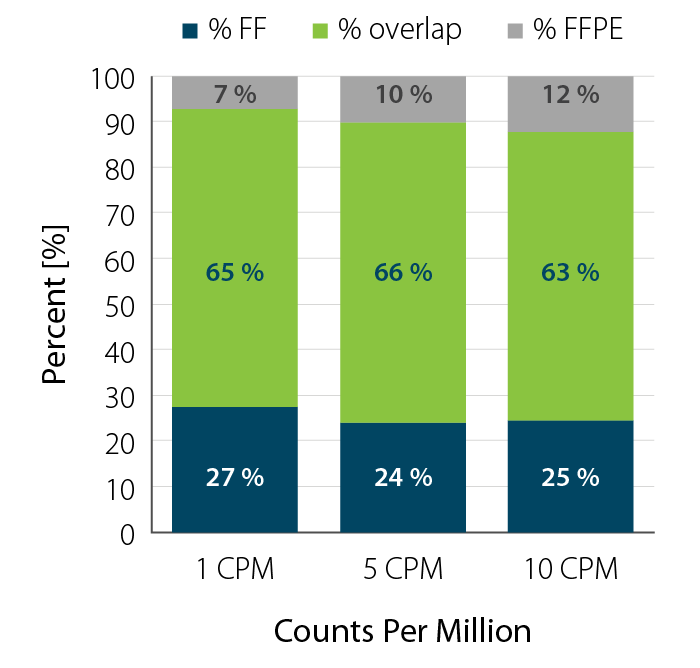
- Degraded RNA (low RIN, high DV200) such as FFPE
- Low inputs (down to 1ng)
System Compatibility:
- iSeq 100
- MiniSeq
- NextSeq 500 – 2000
- HiSeq 2000, 2500, 3000, 4000
- NovaSeq 6000 (v1.0 and 1.5 reagent kits)
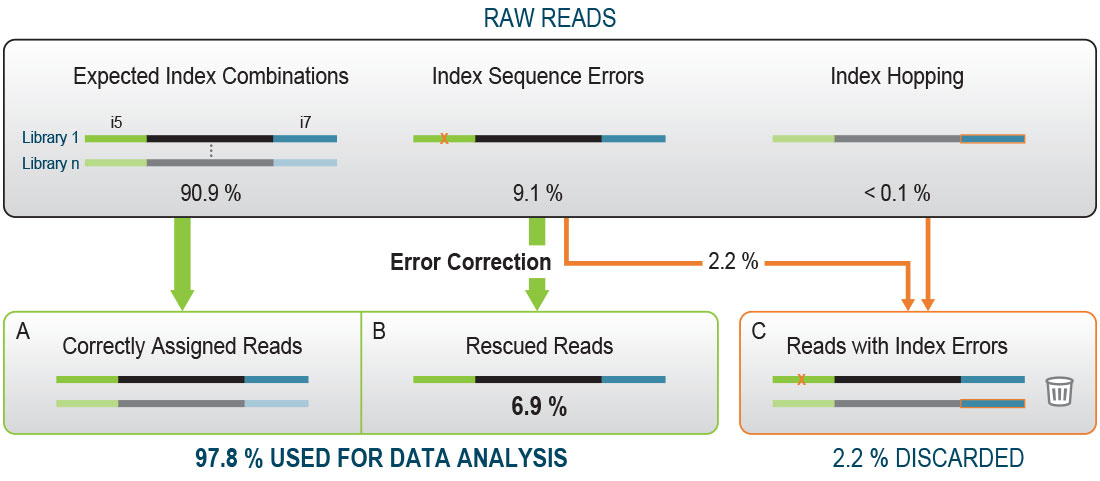
Get the utmost quality out of our well-established QuantSeq 3’ mRNA-Seq gene expression kits, thanks to the improved V2 series! With the increasing reading capacity of next-generation sequencers, Lexogen’s patented 12 nt-long Unique Dual Indices will help you preserve most of your precious reads.
If you already have a library preparation kit without UDI (e.g., QuantSeq-Flex, Reverse, or Forward), our UDI Add-on V2 modules are available for purchase separately. They are delivered together with an improved PCR system, which can be seamlessly adapted to previous QuantSeq kits. For QuantSeq-Pool kits, you only need the UDI Sets (without purchasing additional PCR enzyme and buffer).
Our UDI Add-on V2 kits are also compatible with library preparation kits from other vendors.