Description
NEXTFLEX® Human Core Exome V2 (CES) Panel is designed to overcome the limitations of analyzing genetic variants of interest with respect to conventional whole-exome sequencing.
Sensitive detection of variants of targeted genes of interest.
The NEXTFLEX® Human CES Panel includes significant genes used in other panels and additional genes, therefore allowing customers to obtain comprehensive information of mutations in genes of interest in a cost-effective manner. The 7,513 genes covered in this panel can be reviewed in the NEXTFLEX CES Panel Gene list.
If you are interested in analyzing all of the genes in the human genome, learn about our NEXTFLEX Whole Exome Sequencing (WES) Panel.
Key Features
-
Hard-to-capture regions available with accurate result
-
Comprehensive genomic profile of a variety of genetic disorders
-
Wide range of target regions
-
Automated on Sciclone® G3 NGSx workstation
- Bead-based normalization
Exhaustive Coverage for Each Gene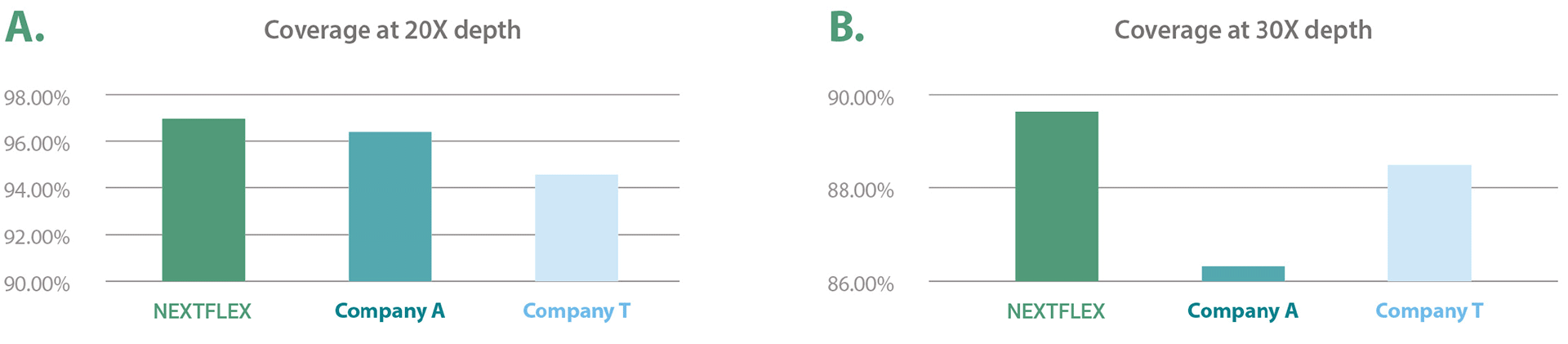 Figure 1: Exhaustive coverage for each gene. The NEXTFLEX CES Panel covers each gene with thorough coverage in comparison to competitor products. The bar graphs indicate the percentage of genes that are covered at (A) 20X depth and (B) 30X depth. The data from the three panels were downsampled to 5.4 Gb.
Figure 1: Exhaustive coverage for each gene. The NEXTFLEX CES Panel covers each gene with thorough coverage in comparison to competitor products. The bar graphs indicate the percentage of genes that are covered at (A) 20X depth and (B) 30X depth. The data from the three panels were downsampled to 5.4 Gb.
Exceptional Target Capture Performance
The NEXTFLEX Human CES Panel provides exceptional target capture performance due to probe design and reagent optimization technology. Despite some companies who resort to masking the hard-to-capture regions (such as GC- or AT-rich regions and homologous regions) or completely omit the regions from their target in order to enhance the quality of their results, the NEXTFLEX Human CES Panel provides both high coverage and on-target ratio without reducing the number of target regions. NEXTFLEX Human CES Panel captures regions that other panels cannot capture with quality coverage and uniformity. The all-around performance of the NEXTFLEX Human CES panel allows for cost-effective and time-saving sequencing of the targeted genes.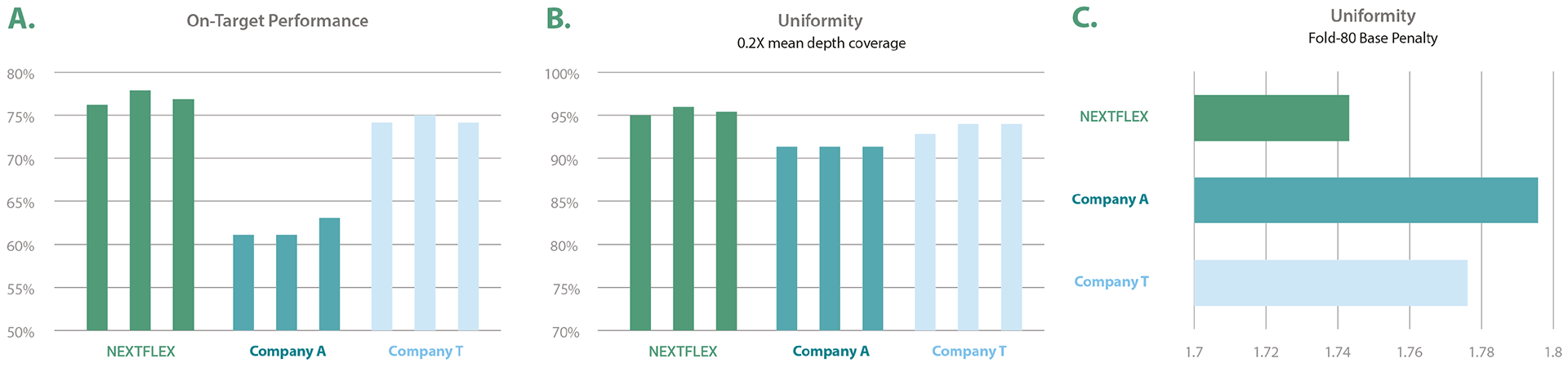 Figure 2: Superior performance in the market. The NEXTFLEX CES Panel shows exceptional performance compared to other competitor products when measured by (A) on-target read ratio, (B) 0.2x mean depth coverage uniformity (higher the better), and (C) Fold-80 base penalty (lower the better). Third-party laboratories conducted a comparison study between the NEXTFLEX CES Panel, Company A, and Company T panels. The same amount of reference materials NA12878, NA12891, and NA12892 were used. Illumina® instruments were used for the sequencing. The data from the three panels were downsampled to 5.4 Gb.
Figure 2: Superior performance in the market. The NEXTFLEX CES Panel shows exceptional performance compared to other competitor products when measured by (A) on-target read ratio, (B) 0.2x mean depth coverage uniformity (higher the better), and (C) Fold-80 base penalty (lower the better). Third-party laboratories conducted a comparison study between the NEXTFLEX CES Panel, Company A, and Company T panels. The same amount of reference materials NA12878, NA12891, and NA12892 were used. Illumina® instruments were used for the sequencing. The data from the three panels were downsampled to 5.4 Gb.
Rapid Same-Day Workflow
Although hybridization capture has great advantages including minimized bias, stable and reliable data results from a variety of sample types, the complexity of the workflow and the long prep time have been obstacles to the users. The NEXTFLEX Human CES Panel workflow significantly simplifies the process and reduces experiment time. Conventional CES takes 2-3 days to complete one sequencing experiment. With NEXTFLEX Human CES Panel workflow, the entire experiment can be completed and the NGS run can be started on the same day.
Streamlined Workflow Reduces Costs & Experiment Time
Instruments such as vacuum concentrators, sonicators, and devices to characterize libraries are barriers against complete automation of core exome sequencing. The NEXTFLEX Human CES panel does not require this instrumentation and alternatively uses enzymes and beads. The incorporation of normalization beads reduces turn-around time by up to three hours mostly due to eliminating the need for individual library quantification prior to the pooling step. This workflow was optimized to enable a complete walkaway solution with reliable performance. The NEXTFLEX Human CES Panel incorporates enzymatic fragmentation, bead-based concentration, and a normalization process, eliminating the need for mechanical fragmentation vacuum concentrator, devices to characterize final libraries.
Full Bioinformatics Capability
Figure 3: Bioinformatics Support. Analysis Service powered by Celemics provides easy data transmission by single-click and automated uploads. It also supports real-time troubleshooting throughout primary and secondary analysis.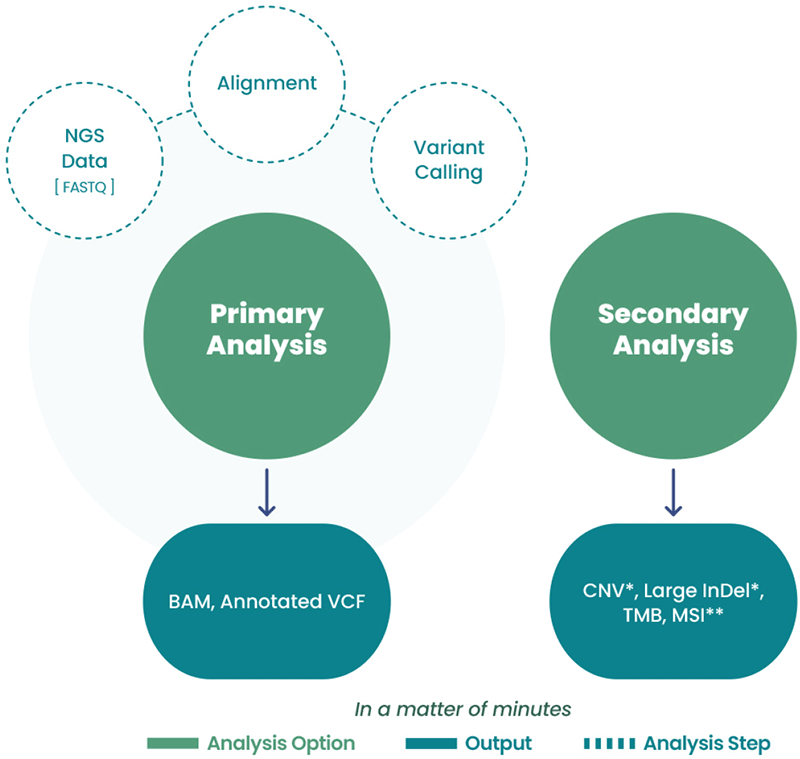
Automation Compatibility
The NEXTFLEX Human CES Panel has been automated on the Sciclone G3 NGSx workstation to increase throughput, minimize sample-to-sample variability and human errors, and reduce hands-on time.
Publications:
- Sabari JK et al. 2017. The activity, safety, and evolving role of brigatinib in patients with ALK-rearranged non-small cell lung cancers. OncoTargets and therapy 10:1983-1992 .
- ASCO_abstr 9007
- ASCO_abstr 9060
- Kim DW et al. 2017. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol. 35 (22):2490-2498 .
- ASCO_abstr e20502
- Gettinger SN et al. 2016. Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: a single-arm, open-label, phase 1/2 trial. Lancet Oncol. 17 (12):1683-1696 .
- Zhang S et al. 2016. The Potent ALK Inhibitor Brigatinib (AP26113) Overcomes Mechanisms of Resistance to Firstand Second-Generation ALK Inhibitors in Preclinical Models. Clin Cancer Res. 22 (22):5527-5538 .
- Kwak EL et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363 (18):1693-703 .
- Soda M et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448 (7153):561-6 .
- Sasaki T et al. 2010. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur. J. Cancer 46 (10):1773-80 .


