Description
CORALL mRNA-Seq V2
Whole transcriptome mRNA sequencing is perfectly suited for expression analysis and delivers gene and transcript level quantification even for low input samples.
Get the best sequencing data with CORALL mRNA-Seq V2. The kit enables fast and cost-efficient generation of stranded, UMI labelled, and unique dual indexed libraries with adjustable insert size.
Performance
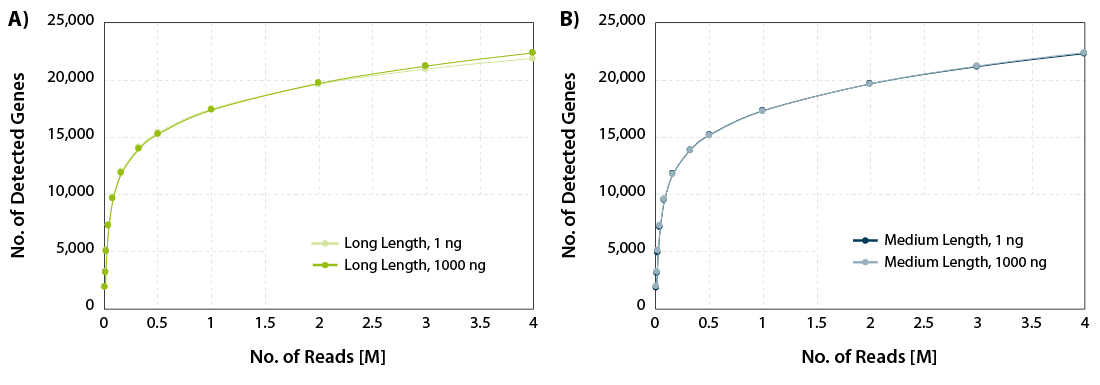
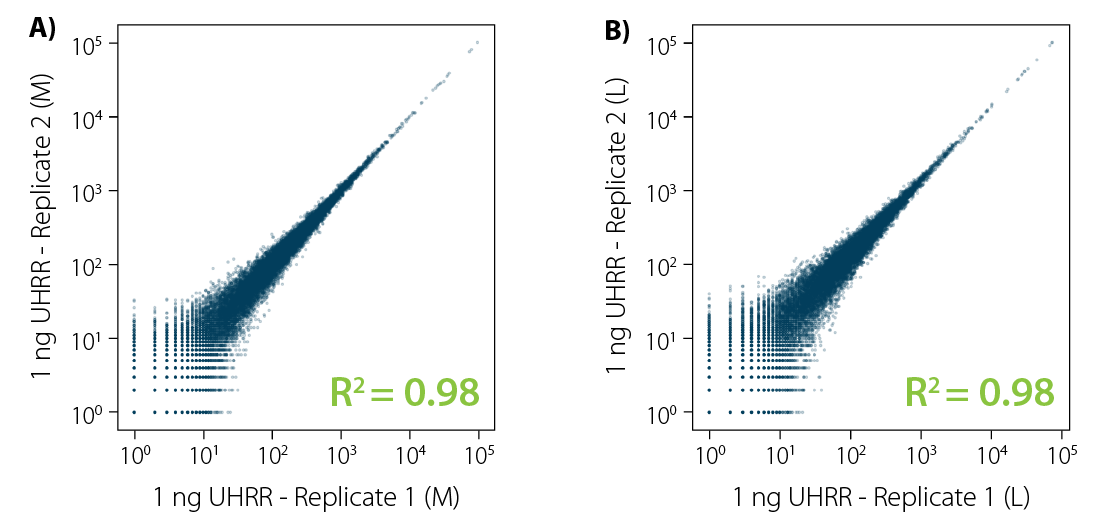
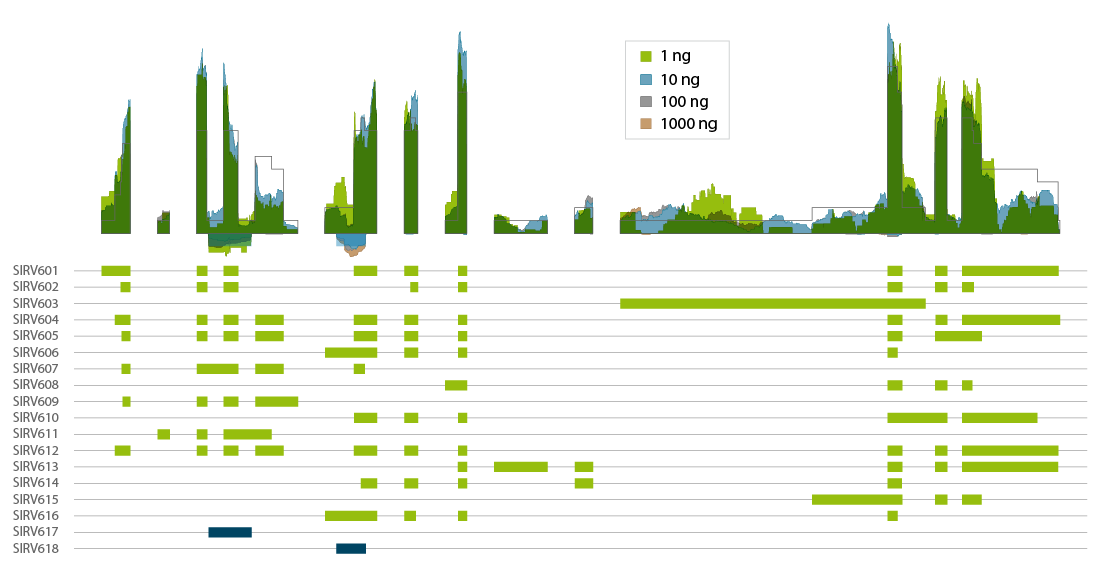
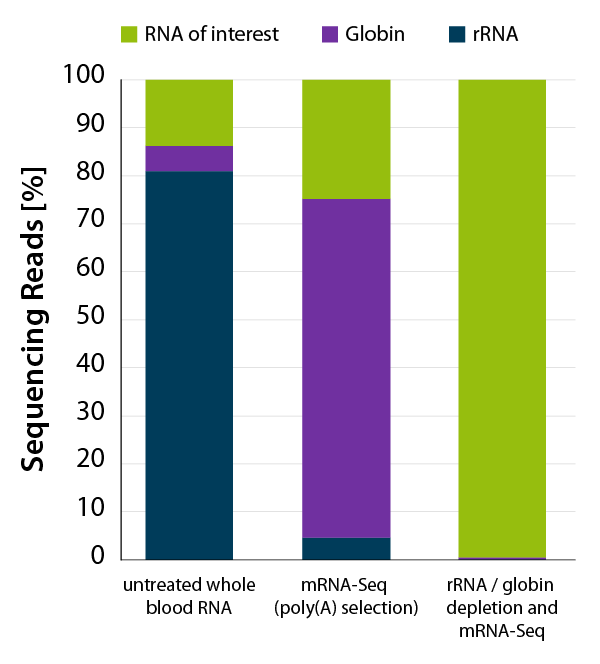
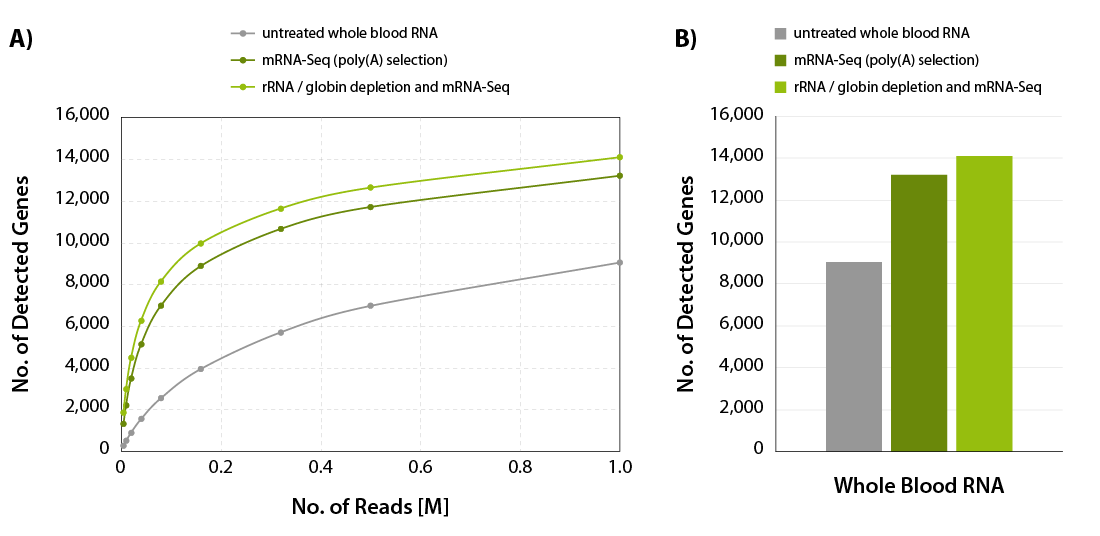
CORALL mRNA-Seq V2 is a fast and flexible whole transcriptome sequencing kit with exceptional performance on low input RNA samples.
Table 1 | CORALL RNA-Seq V2 workflow specifications
| Feature | Specification | Benefit | ||
| CORALL RNA-Seq V2 (stand-alone Kit) | CORALL mRNA-Seq V2 with Poly(A) Selection | CORALL Total RNA-Seq V2 with RiboCop | ||
| Input RNA quantity | 0.1 ng – 100 ng | 1 ng – 1000 ng | 1 ng – 1000 ng | Supports the widest input range available on the market, delivers consistent library output over the full input range |
| RNA types | Total RNA | Poly(A) RNA | Ribo-depleted RNA | Supports all Whole Transcriptome RNA-Seq Applications |
| RNA quality | All qualities, including degraded RNA, FFPE | High-quality RNA (RIN > 8) | All qualities, including degraded RNA, FFPE | Supports all RNA qualities, high sensitivity and high-quality results for difficult material and FFPE samples |
| Workflow time* | 4.5 hours | 5.5 hours | 6 hours | Time-saving workflow with less steps, from sample to sequencing-ready library in one day |
| Kit reaction sizes | 24, 96, 384 | 96, 384 | 24, 96 | Scalable for evaluation, adoption and high-throughout applications |
| Technology | Fragmentation-free library prep using displacement-stop technology | Poly(A) selection by hybridization / capture and CORALL library prep | Hybridization / capture rRNA-depletion without enzymatic digestion and CORALL library prep | – Fragmentation-free workflow – No second strand synthesis – No adaptor titration, less adapter-dimers – Excellent inherent strandedness (> 99 %) |
| Unique Dual Indexing (UDI) | All CORALL RNA-Seq V2 Kits and Bundles contain 12 nt UDI Sets. | Maximized sequencing output on all instruments | ||
| Unique Molecular Identifiers (UMIs) | All CORALL RNA-Seq V2 Kits and Bundles contain built-in UMIs. | Supports identification and removal of PCR duplicates for all sample types and quantities without additional purchases | ||
| Automation Capability | All CORALL RNA-Seq V2 workflows can be automated on liquid handlers. | Supports high-throughput applications | ||
* Workflow time is based upon incubation times and expected times for hands-on-steps. Actual workflow time may vary.
CORALL mRNA-Seq Library Prep Kit
Product Flyer
User Guide for CORALL V2 – update 12.07.2023
User Guide for PCR Add-on and Reamplification Kit V2 – update 08.08.2023
User Guide for Poly(A) Selection Kit – update 13.12.2022
Lexogen UDI 12 nt Unique Dual Index Sequences – update 26.03.2021