Description
The NEXTFLEX® UDI Barcodes (1-1,536) are 1,536 unique dual index adapters with 10 nt indexes for multiplexing of NGS libraries compatible with Illumina® sequencers. They are available off-the-shelf adapters in a set of 1,536 barcodes and in sets of 384 UDI barcodes.
NEXTFLEX® UDI Barcodes (1-1,536) for Ultra High-Throughput Multiplexing
- Sets of 384 UDI barcodes for multiplexing NGS libraries compatible with Illumina® sequencers
- Up to 1,536 (10-nt) adapters available for purchase off-the-shelf
- Every lot is functionally validated and tested for index purity by sequencing
- NEXTFLEX® Universal Blockers compatible with the NEXTFLEX® UDI Barcodes (1-1,536) are also available
| Barcodes |
Barcodes 385-768
|
|---|
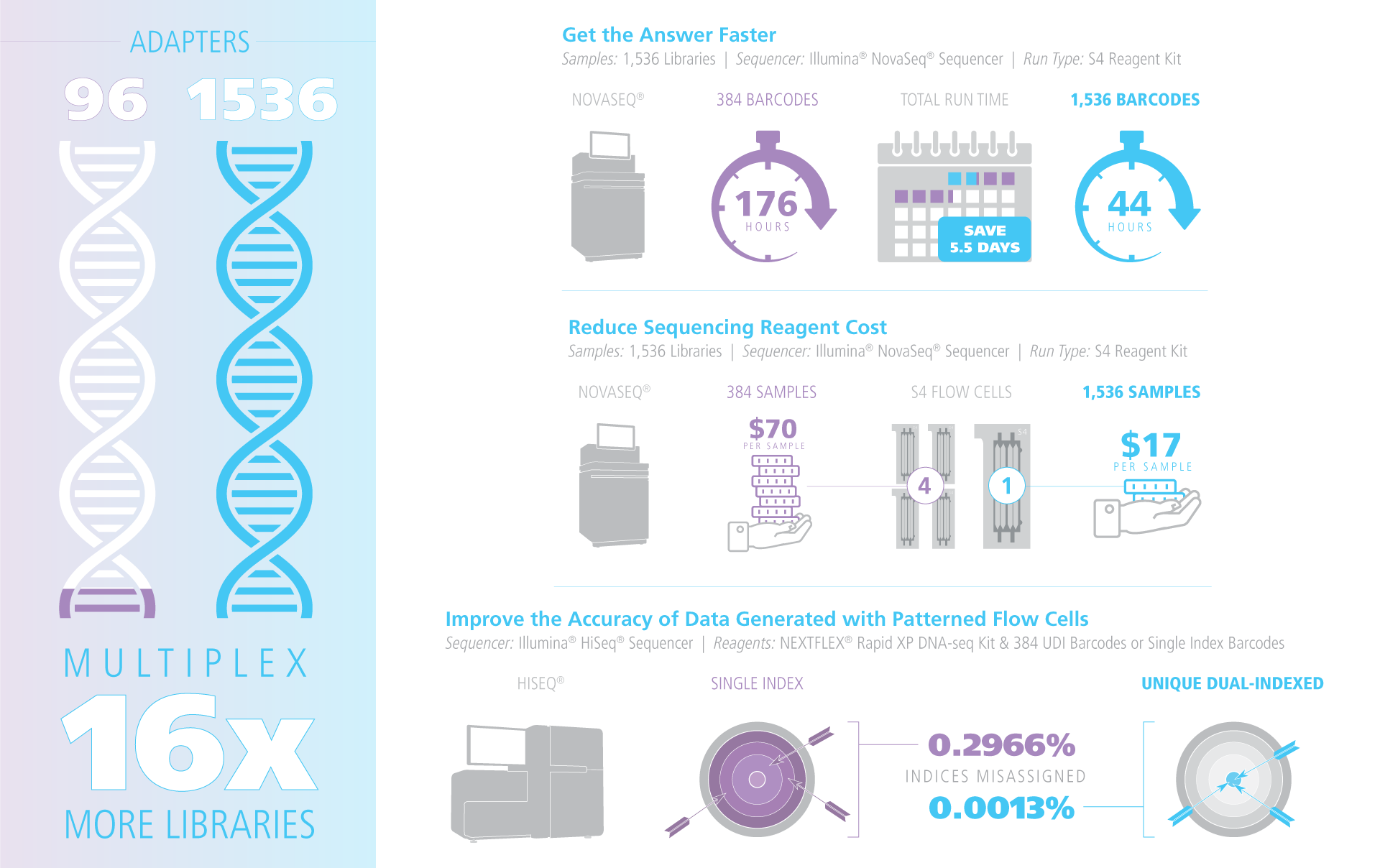
Highest Sample Multiplexing Options for Illumina Sequencers
The design of the NEXTFLEX Unique Dual Index barcodes allow a broad range of multiplexing – users can multiplex anywhere between two samples and 1,536 samples in a single sequencing run. The wide range of multiplexing options enables barcode rotation (avoiding frequent use of the same barcode set) to mitigate run-to-run sample carryover and crosstalk. NEXTFLEX Unique Dual Index barcodes are compatible with NEXTFLEX library prep kits designed for paired-end and single-read sequencing, or other workflows for Illumina sequencers that involve the ligation of adapters to adenylated fragments.
Highest Level of Data Integrity
The NEXTFLEX Unique Dual Index barcodes utilize two unique indexes on each UDI adapter, with a hamming distance of at least three throughout the entire series to permit error correction. Each NEXTFLEX Unique Dual Index barcode comprises the entire flow cell binding site, sequencing primer binding site and index sequences, thereby making PCR enrichment optional. Patterned flow cells, such as those found on the Illumina NovaSeq®, HiSeq® 3000/4000, and HiSeq X platforms, have been known to suffer from increased rates of sample mis-assignment during sequencing. The use of adapters with unique dual indices improves data integrity by correctly removing mis-assigned reads. Every lot is tested for index purity and functionality by sequencing.
 FAQs
FAQs
A: Yes, each lot of the adapters undergoes stringent QC procedures that include verification of the index sequence purity by sequencing on an Illumina MiSeq instrument.
Q: Are the NEXTFLEX unique dual index barcodes functionally validated?
A: Yes, each lot of these adapters undergoes stringent QC procedures that include functional validation of the barcoded adapter by making a library.
Q: What is the lead time when I order the NEXTFLEX unique dual index barcodes?
A: They are in-stock and ready-to-ship.
Q: Why are the NEXTFLEX unique dual index barcodes only supplied at two reactions of each barcode in the 96-well plate?
A: The NEXTFLEX unique dual index barcodes are supplied in aliquots of two reactions of each barcode to reduce the potential for contamination, which increases with repeated use of the same plate.
Q: Must I perform paired-end sequencing with NEXTFLEX Unique Dual Index Barcodes?
A: No, they are compatible with paired-end and single-read sequencing runs. The I7 and I5 indexes are read in separate sequencing events than Read 1 and Read 2.
Q: Can I use the NEXTFLEX Unique Dual Index Barcodes in single index experiments?
A: Yes, you can configure your sequencing run to only read the I7 index when you set up the run.
Selected Citations that Reference the Use of the NEXTFLEX UDI Barcodes:
- Adolfi, A., Gantz, V.M., Jasinskiene, N. et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat Commun 11, 5553 (2020). https://doi.org/10.1038/s41467-020-19426-0
- Estermann, M. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. doi:10.1242/prelights.18906.
- Gardner, E. J., Prigmore, E., Gallone, G., Danecek, P., Samocha, K. E., Handsaker, J., . . . Hurles, M. E. (2019). Contribution of retrotransposition to developmental disorders. Nature Communications, 10(1). doi:10.1038/s41467-019-12520-y.
- Gaeta, N. C., Bean, E., Miles, A. M., Daniel Ubriaco Oliveira Gonçalves De Carvalho, Alemán, M. A., Carvalho, J. S., . . . Ganda, E. (2020). A Cross-Sectional Study of Dairy Cattle Metagenomes Reveals Increased Antimicrobial Resistance in Animals Farmed in a Heavy Metal Contaminated Environment. Frontiers in Microbiology, 11. doi:10.3389/fmicb.2020.590325.
- Hirose, K., Chang, S., Yu, H., Wang, J., Barca, E., Chen, X., . . . Huang, G. N. (2019). Loss of a novel striated muscle-enriched mitochondrial protein Coq10a enhances postnatal cardiac hypertrophic growth. doi:10.1101/755793.
- Leon, K. E., et al. (2020) DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. bioRxiv 2020.08.21.258020; doi: 10.1101/2020.08.21.258020
- Miura, H., Takahashi, S., Shibata, T. et al. Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing. Nat Protoc 15, 4058–4100 (2020). doi.org/10.1038/s41596-020-0378-5
- Starr, T. N., Greaney, A. J., Hilton, S. K., Crawford, K. H., Navarro, M. J., Bowen, J. E., Bloom, J. D. (2020). Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. doi:10.1101/2020.06.17.157982.
- Veenvliet, J. V., et al. (2020) Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. bioRxiv 2020.03.04.974949; doi.org/10.1101/2020.03.04.974949.


