Description
Boost microbial detection with unbiased human DNA depletion.
Human microbiome samples often consist of a mixture of human and microbial DNA. For some samples, contaminating human DNA can be such a large proportion of reads that the sampling of microbial fragments is significantly inhibited. Spending sequencing reads on contaminating human DNA leads to decreased microbial genome coverage, decreased microbial community resolution, and increased sequencing costs. The Human DNA Depletion Kit utilizes a simple CRISPR-based depletion to cut unwanted human molecules that are subsequently removed by an exonuclease treatment. This kit gives you the ability to sequence more microbial reads from your metagenomic samples and get the most comprehensive and unbiased view of the microbiome.
Be among the first to use this early access kit.
Highlights
- Unbiased view of the human microbiome
- Specific CRISPR-mediated depletion of human DNA
- Improved microbial genome coverage previously obscured by human DNA
- Increased detection sensitivity of low abundant microbial species
| Specification | |
| Catalog | JUM-KIT2000 |
|---|---|
| Samples per kit | 24 |
| Assay time | 8 hours |
| Hands-on time | 2.5 hours |
| Sample type | Human-derived tissues with high human DNA contamination |
| Input | 50 ng – 1 ug |
| Input DNA size | It is recommended that prior to starting the depletion protocol to use size selection for larger fragments |
| Designed to deplete | Human DNA fragments |
| validated DNA library prep | Illumina DNA Prep, Tagmentation |
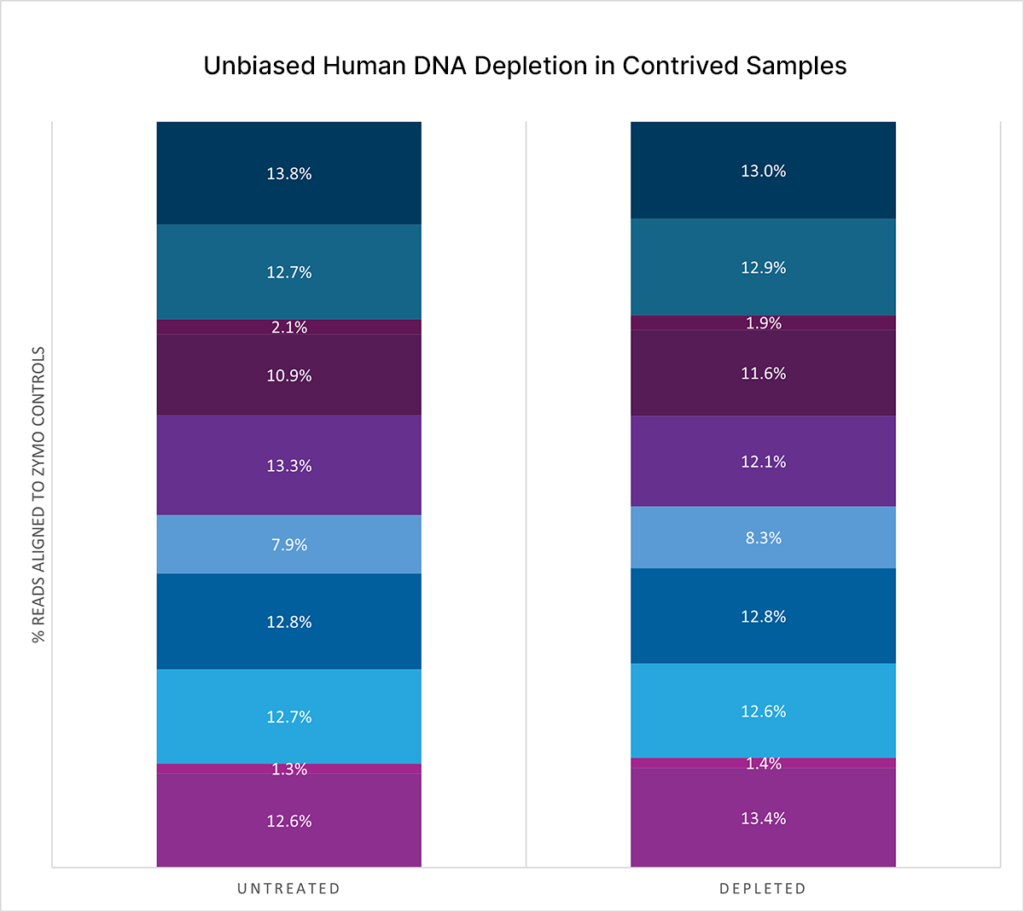
Unbiased human DNA depletion in contrived samples
Boost microbial coverage with less sequencing
Significant increase in breadth of coverage for S. aureus at 10M reads after depletion for contrived samples spiked-in with Zymo community control at 99% human and 1% microbial.

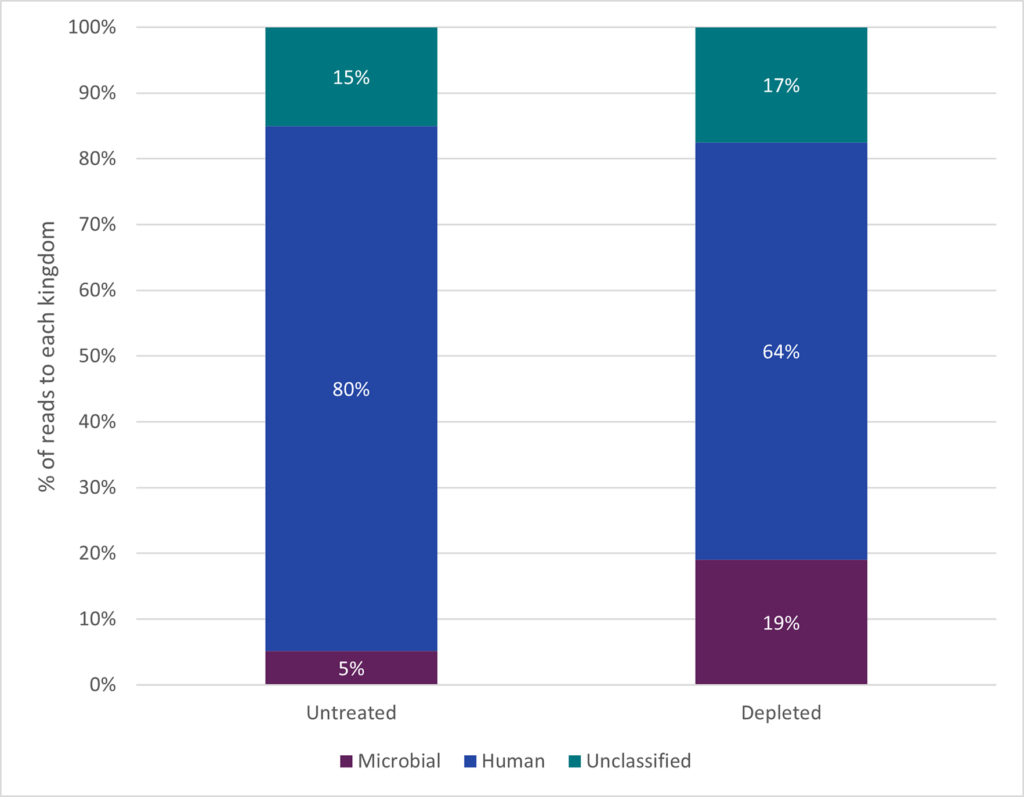
Depletion of human DNA results in read allocation to microbial sequences in clinical vaginal sample.
>10-fold change in reads aligning to microbial sequences found in contrived samples spiked-in with Zymo controls (99% human and 1% microbial).

Improved species detection
Depletion increases the number of species detected from 10 to 46 species in clinical vaginal sample. The original 10 species found in untreated is also found in the depleted clinical vaginal sample.
One day workflow from DNA to library prep ready
8 hours total assay time with 2.5 hours of hands-on time and 2 safe stopping points.