Description
Any Index Set, Made up of: Box 1: Tagging reagent (96w plate) and LPM (Stored at -20°C on arrival), and Box 2: X-solution and 3X coding buffer (Store at ambient temperature on arrival)
Realize the potential of PacBio long read sequencing using seqWell’s LongPlex™ Long Fragment Multiplexing Kit that rapidly multiplexes samples for increased sequencing throughput and cost-effectiveness.
- Plate-based, enzymatic method to simultaneously fragment & index genomic DNA for scalable PacBio HiFi sequencing
- Eliminates the time and cost of mechanical shearing
- Run more long read samples per flow cell without compromising PacBio HiFi sequencing quality
- Supports long read applications including microbial and small genome sequencing, metagenomics, low pass sequencing, and targeted hybrid capture
LongPlex Specifications
| Specs | Description |
|---|---|
| Primary Applications |
Long read sequencing for:
|
| Sample Types* | Genomic DNA (DIN ≥8.0 recommended) |
| Reactions per Kit | 96 reactions |
| DNA Input Recommended | 250 – 500ng |
| Indexing Method | Combinatorial Dual Indexing |
| Number of Unique Index Combinations | 96 |
| Batch Size** | 1-24 samples |
| Output Fragment Range*** | 5,000 – 8,500bp |
| PCR Amplification |
|
| Total Protocol Time |
|
| Sequencer Compatibility | PacBio Revio
PacBio Sequel II & IIe |
*Other sample types may be compatible. Contact seqWell’s support for guidance. **Up to 96 samples can be pooled past LongPlex barcoding prior to PacBio SMRTbell but it may impact read output and coverage uniformity ***Fragment size will depend on magnetic bead cleanup ratios used
LongPlex Long Fragment Multiplexing Kit Includes:
- Tagging Reagent Plate (96-well microtiter plate)
- 3X Coding Buffer
- X Solution
- Library Primary Mix for optional PCR
Key Benefits: Realize the potential of your PacBio long read sequencer
Realize the throughput and cost-effectiveness of PacBio HiFi sequencing using seqWell’s LongPlex Long Fragment Multiplexing Kit. Its speed, simplicity, and scalability enable massive sample multiplexing that unleashes scalable long read sequencing.
- SPEED: Eliminate mechanical shearing using a fully scalable, enzymatic method to simultaneously fragment & tag genomic DNA
- SIMPLICITY: Automation-friendly, plate-based method with total workflow performed in <2 hours (30 minutes hands-on time)
- SCALABILITY: Sample multiplexing using 96 unique dual indexes (UDI) that can be combinatorially expanded with PacBio indexing
- SAVINGS: Early sample pooling greatly reduces all-in cost per sample for long read sequencing without sacrificing data quality
Transposase-based gDNA Fragmentation
Consistent, scalable generation of 8 – 10 kB fragment lengths
Enzymatic fragmentation using adapter-loaded transposase allows for simultaneous fragmentation and indexing of gDNA. Importantly, this eliminates the need for mechanical shearing of gDNA and greatly increases sample throughput.
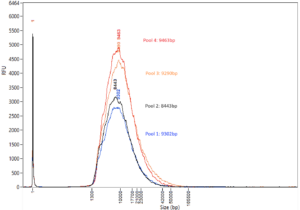
DNA samples were analyzed on the Femto Pulse system following LongPlex processing using the standard PCR-free protocol and SMRTbell 3.0 library preparation. The traces show reproducible fragmentation profiles across four 24-plexes (full plate of 96 samples pooled in 4 separate pools of 24) with an average fragment length >8 kb.
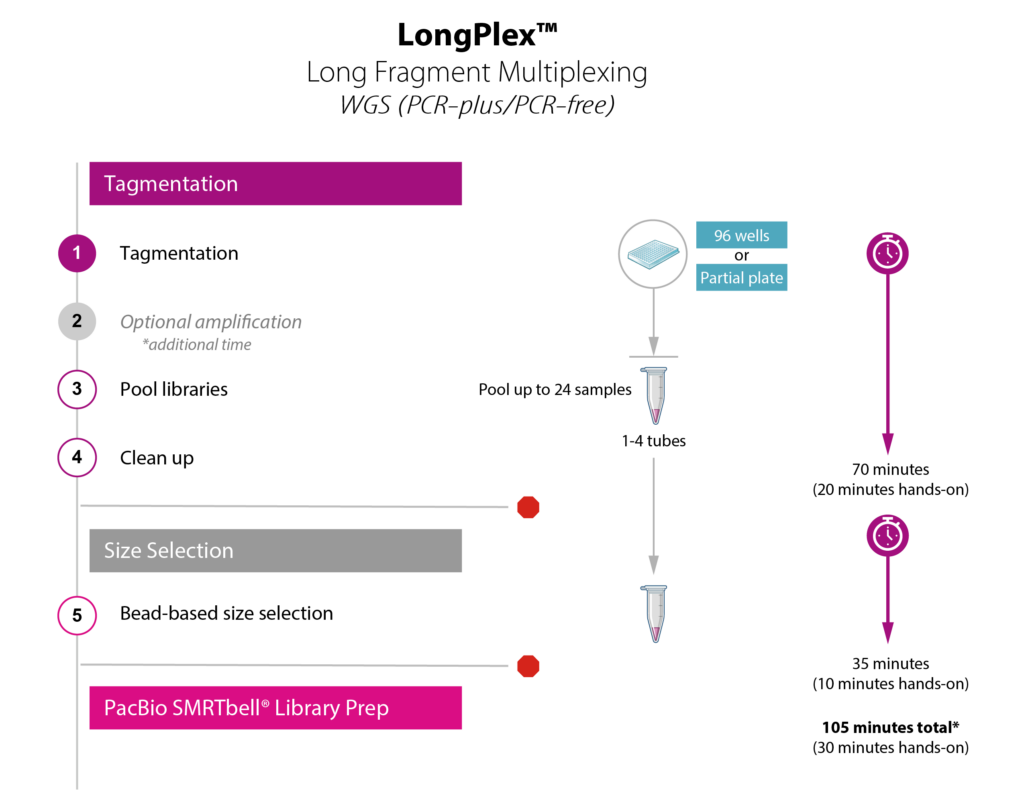
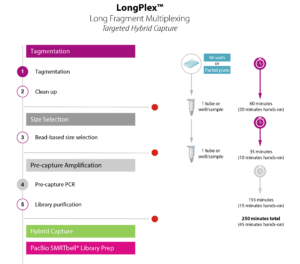
Simple Workflow for Scalable Fragmentation and Multiplexing

- Simultaneously fragment and index up to 96 gDNA samples with no DNA shearing required
- Plate-based protocol performed in < 2 hours with only 30 minutes of hands-on time
- Produces fragments ranging from 8 – 10 kb long
- Sample multiplexing via pooling of up to 96 samples prior to SMRTbell library preparation
- PCR-free and PCR-based protocols available to support a variety of needs including difficult samples
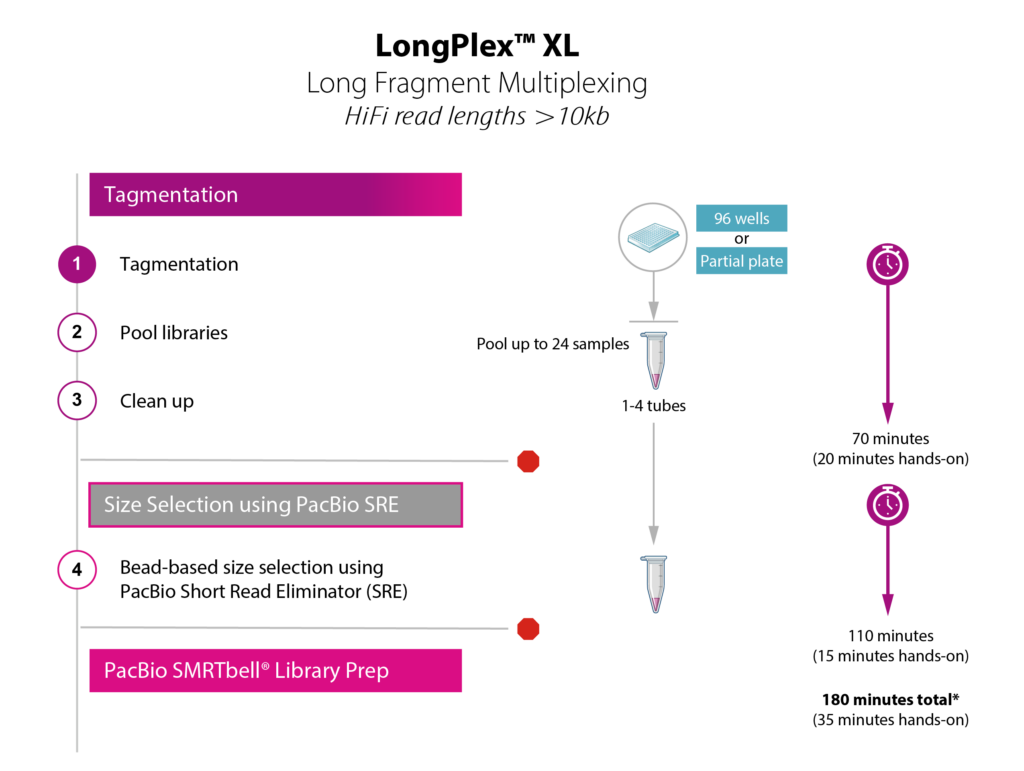
Choose the LongPlex workflow that’s right for you!
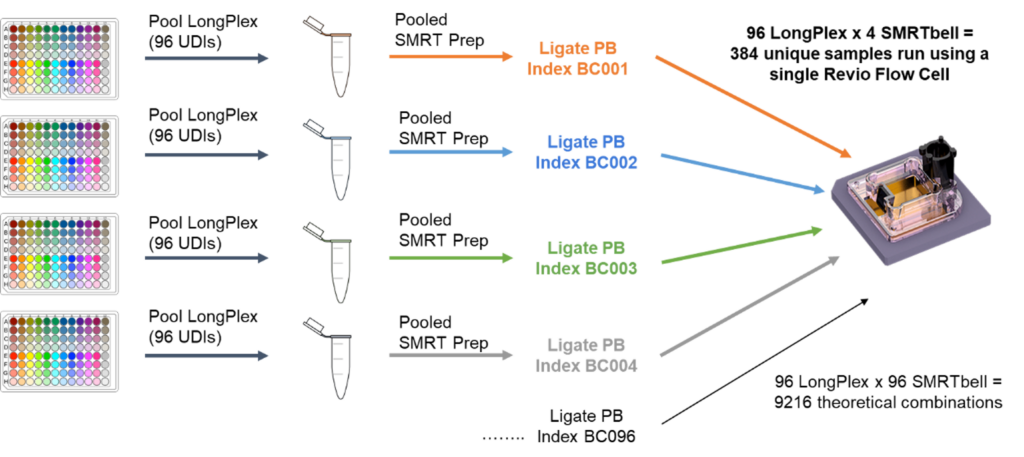
Reducing Long Read Sequencing Costs Through Massive Multiplexing
Efficiently multiplex more samples per flow cell without compromising sequencing quality on PacBio HiFi long-read sequencers. To run 384 long read samples, you no longer need to perform 384 PacBio SMRTbell library preparations! Simply use the plate-based LongPlex method, followed by 4 SMRTbell library preps enabling 384 sample on a single flow cell. Pooling samples early allows you to maximize sequencer throughput while driving down costs.
Downloads: